Serotype-specific differences in dengue virus non-structural protein 5 nuclear localization
- PMID: 23770669
- PMCID: PMC3829348
- DOI: 10.1074/jbc.M113.481382
Serotype-specific differences in dengue virus non-structural protein 5 nuclear localization
Abstract
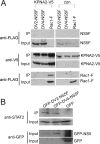
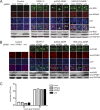
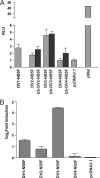
The four serotypes of dengue virus (DENV-1 to -4) cause the most important arthropod-borne viral disease of humans. DENV non-structural protein 5 (NS5) contains enzymatic activities required for capping and replication of the viral RNA genome that occurs in the host cytoplasm. However, previous studies have shown that DENV-2 NS5 accumulates in the nucleus during infection. In this study, we examined the nuclear localization of NS5 for all four DENV serotypes. We demonstrate for the first time that there are serotypic differences in NS5 nuclear localization. Whereas the DENV-2 and -3 proteins accumulate in the nucleus, DENV-1 and -4 NS5 are predominantly if not exclusively localized to the cytoplasm. Comparative studies on the DENV-2 and -4 NS5 proteins revealed that the difference in DENV-4 NS5 nuclear localization was not due to rapid nuclear export but rather the lack of a functional nuclear localization sequence. Interaction studies using DENV-2 and -4 NS5 and human importin-α isoforms failed to identify an interaction that supported the differential nuclear localization of NS5. siRNA knockdown of the human importin-α isoform KPNA2, corresponding to the murine importin-α isoform previously shown to bind to DENV-2 NS5, did not substantially affect DENV-2 NS5 nuclear localization, whereas knockdown of importin-β did. The serotypic differences in NS5 nuclear localization did not correlate with differences in IL-8 gene expression. The results show that NS5 nuclear localization is not strictly required for virus replication but is more likely to have an auxiliary function in the life cycle of specific DENV serotypes.
Keywords: Dengue; Flaviviruses; Nuclear Transport; Positive Strand RNA Viruses; Viral Polymerase; Virology.
Figures




Similar articles
-
Serotype-Specific Regulation of Dengue Virus NS5 Protein Subcellular Localization.ACS Infect Dis. 2024 Jun 14;10(6):2047-2062. doi: 10.1021/acsinfecdis.4c00054. Epub 2024 May 29. ACS Infect Dis. 2024. PMID: 38811007 Free PMC article.
-
Nuclear localization of dengue virus nonstructural protein 5 does not strictly correlate with efficient viral RNA replication and inhibition of type I interferon signaling.J Virol. 2013 Apr;87(8):4545-57. doi: 10.1128/JVI.03083-12. Epub 2013 Feb 13. J Virol. 2013. PMID: 23408610 Free PMC article.
-
Nuclear localization of dengue virus nonstructural protein 5 through its importin alpha/beta-recognized nuclear localization sequences is integral to viral infection.Traffic. 2007 Jul;8(7):795-807. doi: 10.1111/j.1600-0854.2007.00579.x. Epub 2007 May 30. Traffic. 2007. PMID: 17537211
-
The Transactions of NS3 and NS5 in Flaviviral RNA Replication.Adv Exp Med Biol. 2018;1062:147-163. doi: 10.1007/978-981-10-8727-1_11. Adv Exp Med Biol. 2018. PMID: 29845531 Review.
-
Nucleocytoplasmic Trafficking of Dengue Non-structural Protein 5 as a Target for Antivirals.Adv Exp Med Biol. 2018;1062:199-213. doi: 10.1007/978-981-10-8727-1_15. Adv Exp Med Biol. 2018. PMID: 29845535 Review.
Cited by
-
Dengue Virus Non-Structural Protein 5 as a Versatile, Multi-Functional Effector in Host-Pathogen Interactions.Front Cell Infect Microbiol. 2021 Mar 18;11:574067. doi: 10.3389/fcimb.2021.574067. eCollection 2021. Front Cell Infect Microbiol. 2021. PMID: 33816326 Free PMC article. Review.
-
Joint ancestry and association test indicate two distinct pathogenic pathways involved in classical dengue fever and dengue shock syndrome.PLoS Negl Trop Dis. 2018 Feb 15;12(2):e0006202. doi: 10.1371/journal.pntd.0006202. eCollection 2018 Feb. PLoS Negl Trop Dis. 2018. PMID: 29447178 Free PMC article.
-
Nuclear dengue virus NS5 antagonizes expression of PAF1-dependent immune response genes.PLoS Pathog. 2021 Nov 19;17(11):e1010100. doi: 10.1371/journal.ppat.1010100. eCollection 2021 Nov. PLoS Pathog. 2021. PMID: 34797876 Free PMC article.
-
How SARS-CoV-2 and Other Viruses Build an Invasion Route to Hijack the Host Nucleocytoplasmic Trafficking System.Cells. 2021 Jun 7;10(6):1424. doi: 10.3390/cells10061424. Cells. 2021. PMID: 34200500 Free PMC article. Review.
-
High-dimensional CyTOF analysis of dengue virus-infected human DCs reveals distinct viral signatures.JCI Insight. 2017 Jul 6;2(13):e92424. doi: 10.1172/jci.insight.92424. eCollection 2017 Jul 6. JCI Insight. 2017. PMID: 28679950 Free PMC article.
References
-
- World Health Organization (2009) Dengue: Guidelines for Diagnosis, Treatment, Prevention and Control. World Health Organization, Geneva - PubMed
-
- Lindenbach B. D., Thiel H. J., Rice C. M. (2007) Flaviviridae. The viruses and their replication. in Fields Virology, 5th Ed (Knipe D. M., Howley P. M., eds) pp. 1101–1152, Lippincott-Raven Publishers, Philadelphia
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

