Evidence of metabolic switching and implications for food safety from the phenome(s) of Salmonella enterica serovar Typhimurium DT104 cultured at selected points across the pork production food chain
- PMID: 23770904
- PMCID: PMC3754156
- DOI: 10.1128/AEM.01041-13
Evidence of metabolic switching and implications for food safety from the phenome(s) of Salmonella enterica serovar Typhimurium DT104 cultured at selected points across the pork production food chain
Abstract
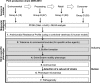
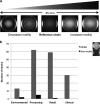
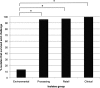
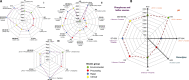
Salmonella enterica serovar Typhimurium DT104 is a recognized food-borne pathogen that displays a multidrug-resistant phenotype and that is associated with systemic infections. At one extreme of the food chain, this bacterium can infect humans, limiting the treatment options available and thereby contributing to increased morbidity and mortality. Although the antibiotic resistance profile is well defined, little is known about other phenotypes that may be expressed by this pathogen at key points across the pork production food chain. In this study, 172 Salmonella enterica serovar Typhimurium DT104/DT104b isolated from an extensive "farm-to-fork" surveillance study, focusing on the pork food chain, were characterized in detail. Isolates were cultured from environmental, processing, retail, and clinical sources, and the study focused on phenotypes that may have contributed to persistence/survival in these different niches. Molecular subtypes, along with antibiotic resistance profiles, tolerance to biocides, motility, and biofilm formation, were determined. As a basis for human infection, acid survival and the ability to utilize a range of energy sources and to adhere to and/or invade Caco-2 cells were also studied. Comparative alterations to biocide tolerance were observed in isolates from retail. l-Tartaric acid and d-mannose-1-phosphate induced the formation of biofilms in a preselected subset of strains, independent of their origin. All clinical isolates were motile and demonstrated an enhanced ability to survive in acidic conditions. Our data report on a diverse phenotype, expressed by S. Typhimurium isolates cultured from the pork production food chain. Extending our understanding of the means by which this pathogen adapts to environmental niches along the "farm-to-fork" continuum will facilitate the protection of vulnerable consumers through targeted improvements in food safety measures.
Figures



Similar articles
-
Microbiological study of biofilm formation in isolates of Salmonella enterica Typhimurium DT104 and DT104b cultured from the modern pork chain.Int J Food Microbiol. 2013 Jan 15;161(1):36-43. doi: 10.1016/j.ijfoodmicro.2012.11.021. Epub 2012 Dec 5. Int J Food Microbiol. 2013. PMID: 23266499
-
Molecular characterization of multidrug-resistant Salmonella enterica subsp. enterica serovar Typhimurium isolates from swine.J Clin Microbiol. 2002 Aug;40(8):2813-22. doi: 10.1128/JCM.40.8.2813-2822.2002. J Clin Microbiol. 2002. PMID: 12149335 Free PMC article.
-
Biofilm formation by multidrug-resistant Salmonella enterica serotype typhimurium phage type DT104 and other pathogens.J Food Prot. 2007 Jan;70(1):22-9. doi: 10.4315/0362-028x-70.1.22. J Food Prot. 2007. PMID: 17265855
-
Non-typhoidal Salmonella in the Pig Production Chain: A Comprehensive Analysis of Its Impact on Human Health.Pathogens. 2019 Jan 29;8(1):19. doi: 10.3390/pathogens8010019. Pathogens. 2019. PMID: 30700039 Free PMC article. Review.
-
Targeting tumors with salmonella Typhimurium- potential for therapy.Oncotarget. 2010 Dec;1(8):721-728. doi: 10.18632/oncotarget.206. Oncotarget. 2010. PMID: 21321381 Free PMC article. Review.
Cited by
-
Salmonella from a Microtidal Estuary Are Capable of Invading Human Intestinal Cell Lines.Microb Ecol. 2020 Feb;79(2):259-270. doi: 10.1007/s00248-019-01419-2. Epub 2019 Aug 5. Microb Ecol. 2020. PMID: 31384980
-
In vivo passage of Salmonella Typhimurium results in minor mutations in the bacterial genome and increases in vitro invasiveness.Vet Res. 2019 Sep 24;50(1):71. doi: 10.1186/s13567-019-0688-1. Vet Res. 2019. PMID: 31551081 Free PMC article.
-
The impact of sseK2 deletion on Salmonella enterica serovar typhimurium virulence in vivo and in vitro.BMC Microbiol. 2019 Aug 7;19(1):182. doi: 10.1186/s12866-019-1543-2. BMC Microbiol. 2019. PMID: 31390974 Free PMC article.
-
Roles of Phosphorus Sources in Microbial Community Assembly for the Removal of Organic Matters and Ammonia in Activated Sludge.Front Microbiol. 2019 May 16;10:1023. doi: 10.3389/fmicb.2019.01023. eCollection 2019. Front Microbiol. 2019. PMID: 31156575 Free PMC article.
-
Responses of Salmonella biofilms to oxidizing biocides: Evidence of spatial clustering.Environ Microbiol. 2022 Dec;24(12):6426-6438. doi: 10.1111/1462-2920.16263. Epub 2022 Nov 6. Environ Microbiol. 2022. PMID: 36300582 Free PMC article.
References
-
- Newell DG, Koopmans M, Verhoef L, Duizer E, Aidara-Kane A, Sprong H, Opsteegh M, Langelaar M, Threfall J, Scheutz F, van der Giessen J, Kruse H. 2010. Food-borne diseases: the challenges of 20 years ago still persist while new ones continue to emerge. Int. J. Food Microbiol. 139:S3–S15 - PMC - PubMed
-
- Zaidi MB, Leon V, Canche C, Perez C, Zhao S, Hubert SK, Abbott J, Blickenstaff K, McDermott PF. 2007. Rapid and widespread dissemination of multidrug-resistant blaCMY-2 Salmonella Typhimurium in Mexico. J. Antimicrob. Chemother. 60:398–401 - PubMed
-
- Alcaine SD, Warnick LD, Wiedmann M. 2007. Antimicrobial resistance in nontyphoidal Salmonella. J. Food Prot. 70:780–790 - PubMed
-
- Davis MA, Hancock DD, Besser TE. 2002. Multiresistant clones of Salmonella enterica: the importance of dissemination. J. Lab. Clin. Med. 140:135–141 - PubMed
-
- Soumpasis I, Butler F. 2009. Development and application of a stochastic epidemic model for the transmission of Salmonella Typhimurium at the farm level of the pork production chain. Risk Anal. 29:1521–1533 - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

