Discovery of plant phenolic compounds that act as type III secretion system inhibitors or inducers of the fire blight pathogen, Erwinia amylovora
- PMID: 23770912
- PMCID: PMC3754148
- DOI: 10.1128/AEM.00845-13
Discovery of plant phenolic compounds that act as type III secretion system inhibitors or inducers of the fire blight pathogen, Erwinia amylovora
Abstract
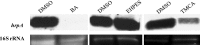
Erwinia amylovora causes a devastating disease called fire blight in rosaceous plants. The type III secretion system (T3SS) is one of the important virulence factors utilized by E. amylovora in order to successfully infect its hosts. By using a green fluorescent protein (GFP) reporter construct combined with a high-throughput flow cytometry assay, a library of phenolic compounds and their derivatives was studied for their ability to alter the expression of the T3SS. Based on the effectiveness of the compounds on the expression of the T3SS pilus, the T3SS inhibitors 4-methoxy-cinnamic acid (TMCA) and benzoic acid (BA) and one T3SS inducer, trans-2-(4-hydroxyphenyl)-ethenylsulfonate (EHPES), were chosen for further study. Both the T3SS inhibitors (TMCA and BA) and the T3SS inducer (EHPES) were found to alter the expression of T3SS through the HrpS-HrpL pathway. Additionally, TMCA altered T3SS expression through the rsmBEa-RsmAEa system. Finally, we found that TMCA and BA weakened the hypersensitive response (HR) in tobacco by suppressing the T3SS of E. amylovora. In our study, we identified phenolic compounds that specifically targeted the T3SS. The T3SS inhibitor may offer an alternative approach to antimicrobial therapy by targeting virulence factors of bacterial pathogens.
Figures


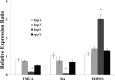


Similar articles
-
Erwinia amylovora Type III Secretion System Inhibitors Reduce Fire Blight Infection Under Field Conditions.Phytopathology. 2023 Dec;113(12):2197-2204. doi: 10.1094/PHYTO-04-23-0111-SA. Epub 2023 Dec 26. Phytopathology. 2023. PMID: 37344783
-
Synthesis and Biological Evaluation of Disulfides Based on Garlic Extract as Type III Secretion System Inhibitors against Erwinia amylovora.J Agric Food Chem. 2025 May 28;73(21):12582-12590. doi: 10.1021/acs.jafc.5c01631. Epub 2025 May 15. J Agric Food Chem. 2025. PMID: 40372403
-
Small-molecule inhibitors suppress the expression of both type III secretion and amylovoran biosynthesis genes in Erwinia amylovora.Mol Plant Pathol. 2014 Jan;15(1):44-57. doi: 10.1111/mpp.12064. Epub 2013 Aug 5. Mol Plant Pathol. 2014. PMID: 23915008 Free PMC article.
-
Virulence Factors of Erwinia amylovora: A Review.Int J Mol Sci. 2015 Jun 5;16(6):12836-54. doi: 10.3390/ijms160612836. Int J Mol Sci. 2015. PMID: 26057748 Free PMC article. Review.
-
Molecular genetics of Erwinia amylovora involved in the development of fire blight.FEMS Microbiol Lett. 2005 Dec 15;253(2):185-92. doi: 10.1016/j.femsle.2005.09.051. Epub 2005 Oct 13. FEMS Microbiol Lett. 2005. PMID: 16253442 Review.
Cited by
-
Polyphenolic extract from maple syrup potentiates antibiotic susceptibility and reduces biofilm formation of pathogenic bacteria.Appl Environ Microbiol. 2015 Jun;81(11):3782-92. doi: 10.1128/AEM.00239-15. Epub 2015 Mar 27. Appl Environ Microbiol. 2015. PMID: 25819960 Free PMC article.
-
Type III secretion inhibitors for the management of bacterial plant diseases.Mol Plant Pathol. 2019 Jan;20(1):20-32. doi: 10.1111/mpp.12736. Epub 2018 Oct 19. Mol Plant Pathol. 2019. PMID: 30062690 Free PMC article.
-
Oleanolic Acid Induces the Type III Secretion System of Ralstonia solanacearum.Front Microbiol. 2015 Dec 22;6:1466. doi: 10.3389/fmicb.2015.01466. eCollection 2015. Front Microbiol. 2015. PMID: 26732647 Free PMC article.
-
Natural Product Type III Secretion System Inhibitors.Antibiotics (Basel). 2019 Sep 24;8(4):162. doi: 10.3390/antibiotics8040162. Antibiotics (Basel). 2019. PMID: 31554164 Free PMC article. Review.
-
Friends and Foes: Bacteria of the Hydroponic Plant Microbiome.Plants (Basel). 2024 Oct 31;13(21):3069. doi: 10.3390/plants13213069. Plants (Basel). 2024. PMID: 39519984 Free PMC article. Review.
References
-
- Norelli JL, Jones AL, Aldwinckle HS. 2003. Fire blight management in the twenty-first century: using new technologies that enhance host resistance in apple. Plant Dis. 87:756–765 - PubMed
-
- Koczan JM, McGrath MJ, Zhao Y, Sundin GW. 2009. Contribution of Erwinia amylovora exopolysaccharides amylovoran and levan to biofilm formation: implications in pathogenicity. Phytopathology 99:1237–1244 - PubMed
-
- Zhang Y, Bak DD, Heid H, Geider K. 1999. Molecular characterization of a protease secreted by Erwinia amylovora. J. Mol. Biol. 289:1239–1251 - PubMed
-
- Dellagi A, Brisset MN, Paulin JP, Expert D. 1998. Dual role of desferrioxamine in Erwinia amylovora pathogenicity. Mol. Plant Microbe Interact. 11:734–742 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

