Sir2 is required for Clr4 to initiate centromeric heterochromatin assembly in fission yeast
- PMID: 23771057
- PMCID: PMC3770337
- DOI: 10.1038/emboj.2013.143
Sir2 is required for Clr4 to initiate centromeric heterochromatin assembly in fission yeast
Abstract
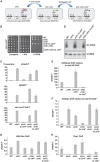
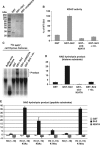
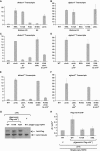
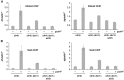
Heterochromatin assembly in fission yeast depends on the Clr4 histone methyltransferase, which targets H3K9. We show that the histone deacetylase Sir2 is required for Clr4 activity at telomeres, but acts redundantly with Clr3 histone deacetylase to maintain centromeric heterochromatin. However, Sir2 is critical for Clr4 function during de novo centromeric heterochromatin assembly. We identified new targets of Sir2 and tested if their deacetylation is necessary for Clr4-mediated heterochromatin establishment. Sir2 preferentially deacetylates H4K16Ac and H3K4Ac, but mutation of these residues to mimic acetylation did not prevent Clr4-mediated heterochromatin establishment. Sir2 also deacetylates H3K9Ac and H3K14Ac. Strains bearing H3K9 or H3K14 mutations exhibit heterochromatin defects. H3K9 mutation blocks Clr4 function, but why H3K14 mutation impacts heterochromatin was not known. Here, we demonstrate that recruitment of Clr4 to centromeres is blocked by mutation of H3K14. We suggest that Sir2 deacetylates H3K14 to target Clr4 to centromeres. Further, we demonstrate that Sir2 is critical for de novo accumulation of H3K9me2 in RNAi-deficient cells. These analyses place Sir2 and H3K14 deacetylation upstream of Clr4 recruitment during heterochromatin assembly.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures



Comment in
-
Histone deacetylases govern heterochromatin in every phase.EMBO J. 2013 Aug 28;32(17):2301-3. doi: 10.1038/emboj.2013.154. Epub 2013 Jul 5. EMBO J. 2013. PMID: 23832177 Free PMC article.
References
-
- Allshire RC, Javerzat JP, Redhead NJ, Cranston G (1994) Position effect variegation at fission yeast centromeres. Cell 76: 157–169 - PubMed
-
- Allshire RC, Nimmo ER, Ekwall K, Javerzat JP, Cranston G (1995) Mutations derepressing silent centromeric domains in fission yeast disrupt chromosome segregation. Genes Dev 9: 218–233 - PubMed
-
- Aparicio OM, Billington BL, Gottschling DE (1991) Modifiers of position effect are shared between telomeric and silent mating-type loci in S. cerevisiae. Cell 66: 1279–1287 - PubMed
-
- Avalos JL, Bever KM, Wolberger C (2005) Mechanism of sirtuin inhibition by nicotinamide: altering the NAD(+) cosubstrate specificity of a Sir2 enzyme. Mol Cell 17: 855–868 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

