Function of PPR proteins in plastid gene expression
- PMID: 23771106
- PMCID: PMC3858428
- DOI: 10.4161/rna.25207
Function of PPR proteins in plastid gene expression
Abstract
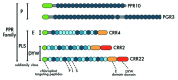
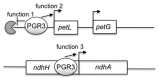
PPR proteins form a huge family in flowering plants and are involved in RNA maturation in plastids and mitochondria. These proteins are sequence-specific RNA-binding proteins that recruit the machinery of RNA processing. We summarize progress in the research on the functional mechanisms of divergent RNA maturation and on the mechanism by which RNA sequences are recognized. We further focus on two topics. RNA editing is an enigmatic process of RNA maturation in organelles, in which members of the PLS subfamily contribute to target site recognition. As the first topic, we speculate on why the PLS subfamily was selected by the RNA editing machinery. Second, we discuss how the regulation of plastid gene expression contributes to efficient photosynthesis. Although the molecular functions of PPR proteins have been studied extensively, information on the physiological significance of regulation by these proteins remains very limited.
Keywords: PPR; RNA; RNA editing; chloroplast; photosynthesis.
Figures

Similar articles
-
Pentatricopeptide repeat proteins involved in plant organellar RNA editing.RNA Biol. 2013;10(9):1419-25. doi: 10.4161/rna.24908. Epub 2013 May 3. RNA Biol. 2013. PMID: 23669716 Free PMC article.
-
RIP1, a member of an Arabidopsis protein family, interacts with the protein RARE1 and broadly affects RNA editing.Proc Natl Acad Sci U S A. 2012 May 29;109(22):E1453-61. doi: 10.1073/pnas.1121465109. Epub 2012 May 7. Proc Natl Acad Sci U S A. 2012. PMID: 22566615 Free PMC article.
-
Seedling Lethal1, a pentatricopeptide repeat protein lacking an E/E+ or DYW domain in Arabidopsis, is involved in plastid gene expression and early chloroplast development.Plant Physiol. 2013 Dec;163(4):1844-58. doi: 10.1104/pp.113.227199. Epub 2013 Oct 21. Plant Physiol. 2013. PMID: 24144791 Free PMC article.
-
RNA editing machinery in plant organelles.Sci China Life Sci. 2018 Feb;61(2):162-169. doi: 10.1007/s11427-017-9170-3. Epub 2017 Oct 23. Sci China Life Sci. 2018. PMID: 29075943 Review.
-
Pentatricopeptide repeat proteins: a socket set for organelle gene expression.Trends Plant Sci. 2008 Dec;13(12):663-70. doi: 10.1016/j.tplants.2008.10.001. Epub 2008 Nov 12. Trends Plant Sci. 2008. PMID: 19004664 Review.
Cited by
-
Temporal aspects of copper homeostasis and its crosstalk with hormones.Front Plant Sci. 2015 Apr 17;6:255. doi: 10.3389/fpls.2015.00255. eCollection 2015. Front Plant Sci. 2015. PMID: 25941529 Free PMC article. Review.
-
The Rice Pentatricopeptide Repeat Protein PPR756 Is Involved in Pollen Development by Affecting Multiple RNA Editing in Mitochondria.Front Plant Sci. 2020 Jun 12;11:749. doi: 10.3389/fpls.2020.00749. eCollection 2020. Front Plant Sci. 2020. PMID: 32595669 Free PMC article.
-
Transcription is a major driving force for plastid genome instability in Arabidopsis.PLoS One. 2019 Apr 3;14(4):e0214552. doi: 10.1371/journal.pone.0214552. eCollection 2019. PLoS One. 2019. PMID: 30943245 Free PMC article.
-
RAP, the sole octotricopeptide repeat protein in Arabidopsis, is required for chloroplast 16S rRNA maturation.Plant Cell. 2014 Feb;26(2):777-87. doi: 10.1105/tpc.114.122853. Epub 2014 Feb 28. Plant Cell. 2014. PMID: 24585838 Free PMC article.
-
Cellular and transcriptomic analyses reveal two-staged chloroplast biogenesis underpinning photosynthesis build-up in the wheat leaf.Genome Biol. 2021 May 11;22(1):151. doi: 10.1186/s13059-021-02366-3. Genome Biol. 2021. PMID: 33975629 Free PMC article.
References
-
- Coffin JW, Dhillon R, Ritzel RG, Nargang FE. The Neurospora crassa cya-5 nuclear gene encodes a protein with a region of homology to the Saccharomyces cerevisiae PET309 protein and is required in a post-transcriptional step for the expression of the mitochondrially encoded COXI protein. Curr Genet. 1997;32:273–80. doi: 10.1007/s002940050277. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
