Dominant Th1 and minimal Th17 skewing in discoid lupus revealed by transcriptomic comparison with psoriasis
- PMID: 23771123
- PMCID: PMC3858414
- DOI: 10.1038/jid.2013.269
Dominant Th1 and minimal Th17 skewing in discoid lupus revealed by transcriptomic comparison with psoriasis
Erratum in
- J Invest Dermatol. 2014 Jun;134(6):1780
Abstract
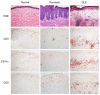
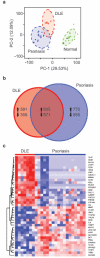
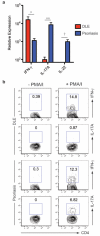
Discoid lupus erythematosus (DLE) is the most common skin manifestation of lupus. Despite its high frequency in systemic lupus in addition to cases without extracutaneous manifestations, targeted treatments for DLE are lacking, likely because of a dearth of knowledge of the molecular landscape of DLE skin. Here, we profiled the transcriptome of DLE skin in order to identify signaling pathways and cellular signatures that may be targeted for treatment purposes. Further comparison of the DLE transcriptome with that of psoriasis, a useful reference given our extensive knowledge of molecular pathways in this disease, provided a framework to identify potential therapeutic targets. Although a growing body of data support a role for IL-17 and T helper type 17 (Th17) cells in systemic lupus, we show a relative enrichment of IFN-γ-associated genes without that for IL-17-associated genes in DLE. Extraction of T cells from the skin of DLE patients identified a predominance of IFN-γ-producing Th1 cells and an absence of IL-17-producing Th17 cells, complementing the results from whole-skin transcriptomic analyses. These data therefore support investigations into treatments for DLE that target Th1 cells or the IFN-γ signaling pathway.
Figures

References
-
- Balanescu P, Balanescu E, Tanasescu C, Nicolau A, Tanasescu R, Grancea C, et al. T helper 17 cell population in lupus erythematosus. Rom J Intern Med. 2010;48:255–9. - PubMed
Publication types
MeSH terms
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

