A role for APETALA1/fruitfull transcription factors in tomato leaf development
- PMID: 23771895
- PMCID: PMC3723613
- DOI: 10.1105/tpc.113.113035
A role for APETALA1/fruitfull transcription factors in tomato leaf development
Abstract
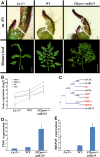
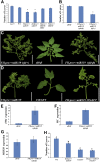
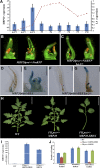
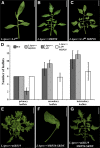
Flexible maturation rates underlie part of the diversity of leaf shape, and tomato (Solanum lycopersicum) leaves are compound due to prolonged organogenic activity of the leaf margin. The CINCINNATA-teosinte branched1, cycloidea, PCF (CIN-TCP) transcription factor lanceolate (LA) restricts this organogenic activity and promotes maturation. Here, we show that tomato APETALA1/fruitfull (AP1/FUL) MADS box genes are involved in tomato leaf development and are repressed by LA. AP1/FUL expression is correlated negatively with LA activity and positively with the organogenic activity of the leaf margin. LA binds to the promoters of the AP1/FUL genes MBP20 and TM4. Overexpression of MBP20 suppressed the simple-leaf phenotype resulting from upregulation of LA activity or from downregulation of class I knotted like homeobox (KNOXI) activity. Overexpression of a dominant-negative form of MBP20 led to leaf simplification and partly suppressed the increased leaf complexity of plants with reduced LA activity or increased KNOXI activity. Tomato plants overexpressing miR319, a negative regulator of several CIN-TCP genes including LA, flower with fewer leaves via an SFT-dependent pathway, suggesting that miR319-sensitive CIN-TCPs delay flowering in tomato. These results identify a role for AP1/FUL genes in vegetative development and show that leaf and plant maturation are regulated via partially independent mechanisms.
Figures


References
-
- Barth S., Geier T., Eimert K., Watillon B., Sangwan R.S., Gleissberg S. (2009). KNOX overexpression in transgenic Kohleria (Gesneriaceae) prolongs the activity of proximal leaf blastozones and drastically alters segment fate. Planta 230: 1081–1091 - PubMed
-
- Becker A., Theissen G. (2003). The major clades of MADS-box genes and their role in the development and evolution of flowering plants. Mol. Phylogenet. Evol. 29: 464–489 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

