Crowders perturb the entropy of RNA energy landscapes to favor folding
- PMID: 23773075
- PMCID: PMC3773054
- DOI: 10.1021/ja4030098
Crowders perturb the entropy of RNA energy landscapes to favor folding
Abstract
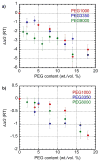
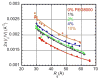
Biological macromolecules have evolved to fold and operate in the crowded environment of the cell. We have shown previously that molecular crowding stabilizes folded RNA structures. Here we report SAXS measurements on a 64 kDa bacterial group I ribozyme in the presence of mono- and divalent ions and PEG crowders of different molecular weight. These experiments show that crowders always stabilize the folded RNA, but this stabilization is weaker in NaCl solutions than MgCl2 solutions. Additionally, we find that RNAs with the same global structure, parametrized by Rg, have different scattering functions depending upon the ratio of electrostatic and entropic stabilization by ions and crowders, respectively. We quantify this difference using the scattering length per scattering volume and find that this ratio is larger for RNAs that fold in lower ionic strength solutions due to the higher crowder content. We conclude that lower RNA flexibility, or reduced configurational entropy, widens the free energy gap between the unfolded and folded RNA in crowded MgCl2 solutions.
Figures





Similar articles
-
Entropic stabilization of folded RNA in crowded solutions measured by SAXS.Nucleic Acids Res. 2016 Nov 2;44(19):9452-9461. doi: 10.1093/nar/gkw597. Epub 2016 Jul 4. Nucleic Acids Res. 2016. PMID: 27378777 Free PMC article.
-
Increased ribozyme activity in crowded solutions.J Biol Chem. 2014 Jan 31;289(5):2972-7. doi: 10.1074/jbc.M113.527861. Epub 2013 Dec 11. J Biol Chem. 2014. PMID: 24337582 Free PMC article.
-
Molecular crowding stabilizes folded RNA structure by the excluded volume effect.J Am Chem Soc. 2010 Jun 30;132(25):8690-6. doi: 10.1021/ja101500g. J Am Chem Soc. 2010. PMID: 20521820 Free PMC article.
-
Molecular crowding and RNA catalysis.Org Biomol Chem. 2020 Oct 14;18(39):7724-7739. doi: 10.1039/d0ob01695k. Org Biomol Chem. 2020. PMID: 32914154 Review.
-
Metal ions and RNA folding: a highly charged topic with a dynamic future.Curr Opin Chem Biol. 2005 Apr;9(2):104-9. doi: 10.1016/j.cbpa.2005.02.004. Curr Opin Chem Biol. 2005. PMID: 15811793 Review.
Cited by
-
Competitive Influence of Alkali Metals in the Ion Atmosphere on Nucleic Acid Duplex Stability.ACS Omega. 2023 Dec 24;9(1):1287-1297. doi: 10.1021/acsomega.3c07563. eCollection 2024 Jan 9. ACS Omega. 2023. PMID: 38222622 Free PMC article.
-
NMR analysis of nucleotide π-stacking in prebiotically relevant crowded environment.Commun Chem. 2020 Apr 30;3(1):51. doi: 10.1038/s42004-020-0300-7. Commun Chem. 2020. PMID: 36703483 Free PMC article.
-
Cellular conditions of weakly chelated magnesium ions strongly promote RNA stability and catalysis.Nat Commun. 2018 Jun 1;9(1):2149. doi: 10.1038/s41467-018-04415-1. Nat Commun. 2018. PMID: 29858572 Free PMC article.
-
Reconstituting Intracellular Vesicle Fusion Reactions: The Essential Role of Macromolecular Crowding.J Am Chem Soc. 2015 Oct 14;137(40):12873-83. doi: 10.1021/jacs.5b08306. Epub 2015 Oct 2. J Am Chem Soc. 2015. PMID: 26431309 Free PMC article.
-
Encapsulation of ribozymes inside model protocells leads to faster evolutionary adaptation.Proc Natl Acad Sci U S A. 2021 May 25;118(21):e2025054118. doi: 10.1073/pnas.2025054118. Proc Natl Acad Sci U S A. 2021. PMID: 34001592 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

