Efficient translation initiation dictates codon usage at gene start
- PMID: 23774758
- PMCID: PMC3964316
- DOI: 10.1038/msb.2013.32
Efficient translation initiation dictates codon usage at gene start
Abstract
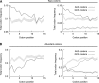
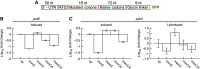
The genetic code is degenerate; thus, protein evolution does not uniquely determine the coding sequence. One of the puzzles in evolutionary genetics is therefore to uncover evolutionary driving forces that result in specific codon choice. In many bacteria, the first 5-10 codons of protein-coding genes are often codons that are less frequently used in the rest of the genome, an effect that has been argued to arise from selection for slowed early elongation to reduce ribosome traffic jams. However, genome analysis across many species has demonstrated that the region shows reduced mRNA folding consistent with pressure for efficient translation initiation. This raises the possibility that unusual codon usage is a side effect of selection for reduced mRNA structure. Here we discriminate between these two competing hypotheses, and show that in bacteria selection favours codons that reduce mRNA folding around the translation start, regardless of whether these codons are frequent or rare. Experiments confirm that primarily mRNA structure, and not codon usage, at the beginning of genes determines the translation rate.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures




References
-
- Beyer D, Skripkin E, Wadzack J, Nierhaus KH (1994) How the ribosome moves along the mRNA during protein synthesis. J Biol Chem 269: 30713–30717 - PubMed
-
- Cannarozzi G, Cannarrozzi G, Schraudolph NN, Faty M, von Rohr P, Friberg MT, Roth AC, Gonnet P, Gonnet G, Barral Y (2010) A role for codon order in translation dynamics. Cell 141: 355–367 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

