Archaea on human skin
- PMID: 23776475
- PMCID: PMC3680501
- DOI: 10.1371/journal.pone.0065388
Archaea on human skin
Abstract
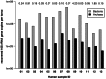
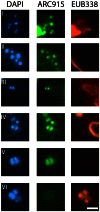
The recent era of exploring the human microbiome has provided valuable information on microbial inhabitants, beneficials and pathogens. Screening efforts based on DNA sequencing identified thousands of bacterial lineages associated with human skin but provided only incomplete and crude information on Archaea. Here, we report for the first time the quantification and visualization of Archaea from human skin. Based on 16 S rRNA gene copies Archaea comprised up to 4.2% of the prokaryotic skin microbiome. Most of the gene signatures analyzed belonged to the Thaumarchaeota, a group of Archaea we also found in hospitals and clean room facilities. The metabolic potential for ammonia oxidation of the skin-associated Archaea was supported by the successful detection of thaumarchaeal amoA genes in human skin samples. However, the activity and possible interaction with human epithelial cells of these associated Archaea remains an open question. Nevertheless, in this study we provide evidence that Archaea are part of the human skin microbiome and discuss their potential for ammonia turnover on human skin.
Conflict of interest statement
Figures

Similar articles
-
Thaumarchaeal ammonia oxidation in an acidic forest peat soil is not influenced by ammonium amendment.Appl Environ Microbiol. 2010 Nov;76(22):7626-34. doi: 10.1128/AEM.00595-10. Epub 2010 Oct 1. Appl Environ Microbiol. 2010. PMID: 20889787 Free PMC article.
-
Aquarium nitrification revisited: Thaumarchaeota are the dominant ammonia oxidizers in freshwater aquarium biofilters.PLoS One. 2011;6(8):e23281. doi: 10.1371/journal.pone.0023281. Epub 2011 Aug 16. PLoS One. 2011. PMID: 21858055 Free PMC article.
-
Niche specialization of terrestrial archaeal ammonia oxidizers.Proc Natl Acad Sci U S A. 2011 Dec 27;108(52):21206-11. doi: 10.1073/pnas.1109000108. Epub 2011 Dec 8. Proc Natl Acad Sci U S A. 2011. PMID: 22158986 Free PMC article.
-
The Thaumarchaeota: an emerging view of their phylogeny and ecophysiology.Curr Opin Microbiol. 2011 Jun;14(3):300-6. doi: 10.1016/j.mib.2011.04.007. Epub 2011 May 4. Curr Opin Microbiol. 2011. PMID: 21546306 Free PMC article. Review.
-
The Archaea domain: Exploring historical and contemporary perspectives with in silico primer coverage analysis for future research in Dentistry.Arch Oral Biol. 2024 May;161:105936. doi: 10.1016/j.archoralbio.2024.105936. Epub 2024 Feb 23. Arch Oral Biol. 2024. PMID: 38422909 Review.
Cited by
-
The Role of Methanogenic Archaea in Inflammatory Bowel Disease-A Review.J Pers Med. 2024 Feb 10;14(2):196. doi: 10.3390/jpm14020196. J Pers Med. 2024. PMID: 38392629 Free PMC article. Review.
-
Cross-Domain and Viral Interactions in the Microbiome.Microbiol Mol Biol Rev. 2019 Jan 9;83(1):e00044-18. doi: 10.1128/MMBR.00044-18. Print 2019 Mar. Microbiol Mol Biol Rev. 2019. PMID: 30626617 Free PMC article. Review.
-
Review article: inhibition of methanogenic archaea by statins as a targeted management strategy for constipation and related disorders.Aliment Pharmacol Ther. 2016 Jan;43(2):197-212. doi: 10.1111/apt.13469. Epub 2015 Nov 11. Aliment Pharmacol Ther. 2016. PMID: 26559904 Free PMC article. Review.
-
Cultivation and characterization of Candidatus Nitrosocosmicus exaquare, an ammonia-oxidizing archaeon from a municipal wastewater treatment system.ISME J. 2017 May;11(5):1142-1157. doi: 10.1038/ismej.2016.192. Epub 2017 Feb 14. ISME J. 2017. PMID: 28195581 Free PMC article.
-
Human Milk Oligosaccharides Modulate the Risk for Preterm Birth in a Microbiome-Dependent and -Independent Manner.mSystems. 2020 Jun 9;5(3):e00334-20. doi: 10.1128/mSystems.00334-20. mSystems. 2020. PMID: 32518196 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

