Distinct microRNAs expression profile in primary biliary cirrhosis and evaluation of miR 505-3p and miR197-3p as novel biomarkers
- PMID: 23776611
- PMCID: PMC3680413
- DOI: 10.1371/journal.pone.0066086
Distinct microRNAs expression profile in primary biliary cirrhosis and evaluation of miR 505-3p and miR197-3p as novel biomarkers
Abstract
Background and aims: MicroRNAs are small endogenous RNA molecules with specific expression patterns that can serve as biomarkers for numerous diseases. However, little is known about the expression profile of serum miRNAs in PBC.
Methods: First, we employed Illumina deep sequencing for the initial screening to indicate the read numbers of miRNA expression in 10 PBC, 5 CH-C, 5 CH-B patients and 5 healthy controls. Comparing the differentially expressed miRNAs in the 4 groups, analysis of variance was performed on the number of sequence reads to evaluate the statistical significance. Hierarchical clustering was performed using an R platform and we have found candidates for specific miRNAs in the PBC patients. Second, a quantitative reverse transcription PCR validation study was conducted in 10 samples in each group. The expression levels of the selected miRNAs were presented as fold-changes (2(-ΔΔCt)). Finally, computer analysis was conducted to predict target genes and biological functions with MiRror 2.0 and DAVID v6.7.
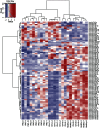
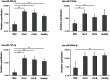
Results: We obtained about 12 million 32-mer short RNA reads on average per sample and the mapping rates to miRBase were 16.60% and 81.66% to hg19. In the statistical significance testing, the expression levels of 81 miRNAs were found to be differentially expressed in the 4 groups. The heat map and hierarchical clustering demonstrated that the miRNA profiles from PBC clustered with those of CH-B, CH-C and healthy controls. Additionally, the circulating levels of hsa-miR-505-3p, 197-3p, and 500a-3p were significantly decreased in PBC compared with healthy controls and the expression levels of hsa-miR-505-3p, 139-5p and 197-3p were significantly reduced compared with the viral hepatitis group.
Conclusions: Our results indicate that sera from patients with PBC have a unique miRNA expression profile and that the down-regulated expression of hsa-miR-505-3p and miR-197-3p can serve as clinical biomarkers of PBC.
Conflict of interest statement
Figures
Similar articles
-
The expression of miR-125b-5p is increased in the serum of patients with chronic hepatitis B infection and inhibits the detection of hepatitis B virus surface antigen.J Viral Hepat. 2016 May;23(5):330-9. doi: 10.1111/jvh.12522. Epub 2016 Feb 29. J Viral Hepat. 2016. PMID: 26924666
-
MicroRNA-223 and microRNA-21 in peripheral blood B cells associated with progression of primary biliary cholangitis patients.PLoS One. 2017 Sep 8;12(9):e0184292. doi: 10.1371/journal.pone.0184292. eCollection 2017. PLoS One. 2017. PMID: 28886078 Free PMC article.
-
Serum microRNAs as potential biomarkers of primary biliary cirrhosis.PLoS One. 2014 Oct 27;9(10):e111424. doi: 10.1371/journal.pone.0111424. eCollection 2014. PLoS One. 2014. PMID: 25347847 Free PMC article.
-
Differential microRNA expression in the peripheral blood from human patients with COVID-19.J Clin Lab Anal. 2020 Oct;34(10):e23590. doi: 10.1002/jcla.23590. Epub 2020 Sep 22. J Clin Lab Anal. 2020. PMID: 32960473 Free PMC article.
-
MiR-139-5p is associated with inflammatory regulation through c-FOS suppression, and contributes to the progression of primary biliary cholangitis.Lab Invest. 2016 Nov;96(11):1165-1177. doi: 10.1038/labinvest.2016.95. Epub 2016 Sep 26. Lab Invest. 2016. PMID: 27668889
Cited by
-
LINC00312 inhibits the migration and invasion of bladder cancer cells by targeting miR-197-3p.Tumour Biol. 2016 Nov;37(11):14553-14563. doi: 10.1007/s13277-016-5303-8. Epub 2016 Sep 8. Tumour Biol. 2016. PMID: 27631965
-
Role of circulating microRNAs in liver diseases.World J Hepatol. 2017 Apr 28;9(12):586-594. doi: 10.4254/wjh.v9.i12.586. World J Hepatol. 2017. PMID: 28515844 Free PMC article. Review.
-
MicroRNAs in the Cholangiopathies: Pathogenesis, Diagnosis, and Treatment.J Clin Med. 2015 Aug 26;4(9):1688-712. doi: 10.3390/jcm4091688. J Clin Med. 2015. PMID: 26343736 Free PMC article. Review.
-
Diagnostic and therapeutic potentials of microRNAs in cholangiopathies.Liver Res. 2017 Jun;1(1):34-41. doi: 10.1016/j.livres.2017.03.003. Epub 2017 Apr 26. Liver Res. 2017. PMID: 29085701 Free PMC article.
-
MiR-197-3p affects angiogenesis and inflammation of endothelial cells by targeting CXCR2/COX2 axis.Am J Transl Res. 2022 Jul 15;14(7):4666-4677. eCollection 2022. Am J Transl Res. 2022. PMID: 35958438 Free PMC article.
References
-
- Lau NC, Lim LP, Weinstein EG, Bartel DP (2001) An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science 294: 858–862. - PubMed
-
- Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T (2001) Identification of novel genes coding for small expressed RNAs. Science 294: 853–858. - PubMed
-
- Lee RC, Ambros V (2001) An extensive class of small RNAs in Caenorhabditis elegans. Science 294: 862–864. - PubMed
-
- Bartel DP (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116: 281–297. - PubMed
-
- Chen X, Ba Y, Ma L, Cai X, Yin Y, et al. (2008) Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Research 18: 997–1006. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

