The complete mitochondrial genome of Solemya velum (Mollusca: Bivalvia) and its relationships with conchifera
- PMID: 23777315
- PMCID: PMC3704766
- DOI: 10.1186/1471-2164-14-409
The complete mitochondrial genome of Solemya velum (Mollusca: Bivalvia) and its relationships with conchifera
Abstract
Background: Bivalve mitochondrial genomes exhibit a wide array of uncommon features, like extensive gene rearrangements, large sizes, and unusual ways of inheritance. Species pertaining to the order Solemyida (subclass Opponobranchia) show many peculiar evolutionary adaptations, f.i. extensive symbiosis with chemoautotrophic bacteria. Despite Opponobranchia are central in bivalve phylogeny, being considered the sister group of all Autobranchia, a complete mitochondrial genome has not been sequenced yet.
Results: In this paper, we characterized the complete mitochondrial genome of the Atlantic awning clam Solemya velum: A-T content, gene arrangement and other features are more similar to putative ancestral mollusks than to other bivalves. Two supranumerary open reading frames are present in a large, otherwise unassigned, region, while the origin of replication could be located in a region upstream to the cox3 gene.
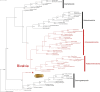
Conclusions: We show that S. velum mitogenome retains most of the ancestral conchiferan features, which is unusual among bivalve mollusks, and we discuss main peculiarities of this first example of an organellar genome coming from the subclass Opponobranchia. Mitochondrial genomes of Solemya (for bivalves) and Haliotis (for gastropods) seem to retain the original condition of mollusks, as most probably exemplified by Katharina.
Figures



References
-
- Simison WB, Boore JL. In: Phylogeny and evolution of the mollusca. Ponder W, Lindberg DR, editor. Berkeley: University of California Press; 2008. Molluscan evolutionary genomics; pp. 447–461.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

