CD4⁺ follicular helper T cell infiltration predicts breast cancer survival
- PMID: 23778140
- PMCID: PMC3696556
- DOI: 10.1172/JCI67428
CD4⁺ follicular helper T cell infiltration predicts breast cancer survival
Abstract
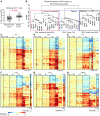
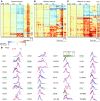
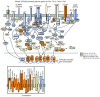
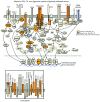
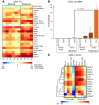
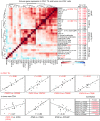
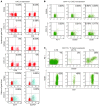
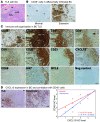
CD4⁺ T cells are critical regulators of immune responses, but their functional role in human breast cancer is relatively unknown. The goal of this study was to produce an image of CD4⁺ T cells infiltrating breast tumors using limited ex vivo manipulation to better understand the in vivo differences associated with patient prognosis. We performed comprehensive molecular profiling of infiltrating CD4⁺ T cells isolated from untreated invasive primary tumors and found that the infiltrating T cell subpopulations included follicular helper T (Tfh) cells, which have not previously been found in solid tumors, as well as Th1, Th2, and Th17 effector memory cells and Tregs. T cell signaling pathway alterations included a mixture of activation and suppression characterized by restricted cytokine/chemokine production, which inversely paralleled lymphoid infiltration levels and could be reproduced in activated donor CD4⁺ T cells treated with primary tumor supernatant. A comparison of extensively versus minimally infiltrated tumors showed that CXCL13-producing CD4⁺ Tfh cells distinguish extensive immune infiltrates, principally located in tertiary lymphoid structure germinal centers. An 8-gene Tfh signature, signifying organized antitumor immunity, robustly predicted survival or preoperative response to chemotherapy. Our identification of CD4⁺ Tfh cells in breast cancer suggests that they are an important immune element whose presence in the tumor is a prognostic factor.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

