Functions of cofilin in cell locomotion and invasion
- PMID: 23778968
- PMCID: PMC3878614
- DOI: 10.1038/nrm3609
Functions of cofilin in cell locomotion and invasion
Abstract
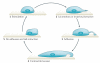
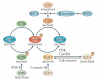
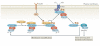
Recently, a consensus has emerged that cofilin severing activity can generate free actin filament ends that are accessible for F-actin polymerization and depolymerization without changing the rate of G-actin association and dissociation at either filament end. The structural basis of actin filament severing by cofilin is now better understood. These results have been integrated with recently discovered mechanisms for cofilin activation in migrating cells, which led to new models for cofilin function that provide insights into how cofilin regulation determines the temporal and spatial control of cell behaviour.
Figures



References
-
- Charras G, Paluch E. Blebs lead the way: how to migrate without lamellipodia. Nature Rev. Mol. Cell Biol. 2008;9:730–736. - PubMed
-
- Daubon T, Rochelle T, Bourmeyster N, Genot E. Invadopodia and rolling-type motility are specific features of highly invasive p190bcr-abl leukemic cells. Eur. J. Cell Biol. 2012;91:978–987. - PubMed
-
- Nurnberg A, Kitzing T, Grosse R. Nucleating actin for invasion. Nature Rev. Cancer. 2011;11:177–187. - PubMed
-
- Ridley AJ. Life at the leading edge. Cell. 2011;145:1012–1022. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

