Distinct evolutionary trajectories of primary high-grade serous ovarian cancers revealed through spatial mutational profiling
- PMID: 23780408
- PMCID: PMC3864404
- DOI: 10.1002/path.4230
Distinct evolutionary trajectories of primary high-grade serous ovarian cancers revealed through spatial mutational profiling
Abstract
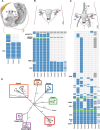
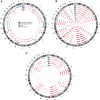
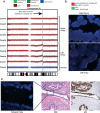
High-grade serous ovarian cancer (HGSC) is characterized by poor outcome, often attributed to the emergence of treatment-resistant subclones. We sought to measure the degree of genomic diversity within primary, untreated HGSCs to examine the natural state of tumour evolution prior to therapy. We performed exome sequencing, copy number analysis, targeted amplicon deep sequencing and gene expression profiling on 31 spatially and temporally separated HGSC tumour specimens (six patients), including ovarian masses, distant metastases and fallopian tube lesions. We found widespread intratumoural variation in mutation, copy number and gene expression profiles, with key driver alterations in genes present in only a subset of samples (eg PIK3CA, CTNNB1, NF1). On average, only 51.5% of mutations were present in every sample of a given case (range 10.2-91.4%), with TP53 as the only somatic mutation consistently present in all samples. Complex segmental aneuploidies, such as whole-genome doubling, were present in a subset of samples from the same individual, with divergent copy number changes segregating independently of point mutation acquisition. Reconstruction of evolutionary histories showed one patient with mixed HGSC and endometrioid histology, with common aetiologic origin in the fallopian tube and subsequent selection of different driver mutations in the histologically distinct samples. In this patient, we observed mixed cell populations in the early fallopian tube lesion, indicating that diversity arises at early stages of tumourigenesis. Our results revealed that HGSCs exhibit highly individual evolutionary trajectories and diverse genomic tapestries prior to therapy, exposing an essential biological characteristic to inform future design of personalized therapeutic solutions and investigation of drug-resistance mechanisms.
Keywords: clonal evolution; high-grade serous ovarian cancer; intratumoural heterogeneity.
© 2013 The Authors. Journal of Pathology published by John Wiley & Sons Ltd on behalf of Pathological Society of Great Britain and Ireland.
Figures




References
-
- Piccart MJ, Bertelsen K, James K, et al. Randomized intergroup trial of cisplatin–paclitaxel versus cisplatin–cyclophosphamide in women with advanced epithelial ovarian cancer: three-year results. J Natl Cancer Inst. 2000;92::699–708. - PubMed
-
- Risch HA, McLaughlin JR, Cole DEC, et al. Population BRCA1 and BRCA2 mutation frequencies and cancer penetrances: a kin–cohort study in Ontario, Canada. J Natl Cancer Inst. 2006;98::1694–1706. - PubMed
-
- Geisler JP, Hatterman-Zogg MA, Rathe JA, et al. Frequency of BRCA1 dysfunction in ovarian cancer. J Natl Cancer Inst. 2002;94::61–67. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

