The Virtual Brain: a simulator of primate brain network dynamics
- PMID: 23781198
- PMCID: PMC3678125
- DOI: 10.3389/fninf.2013.00010
The Virtual Brain: a simulator of primate brain network dynamics
Abstract
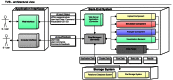
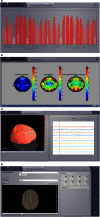
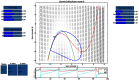
We present The Virtual Brain (TVB), a neuroinformatics platform for full brain network simulations using biologically realistic connectivity. This simulation environment enables the model-based inference of neurophysiological mechanisms across different brain scales that underlie the generation of macroscopic neuroimaging signals including functional MRI (fMRI), EEG and MEG. Researchers from different backgrounds can benefit from an integrative software platform including a supporting framework for data management (generation, organization, storage, integration and sharing) and a simulation core written in Python. TVB allows the reproduction and evaluation of personalized configurations of the brain by using individual subject data. This personalization facilitates an exploration of the consequences of pathological changes in the system, permitting to investigate potential ways to counteract such unfavorable processes. The architecture of TVB supports interaction with MATLAB packages, for example, the well known Brain Connectivity Toolbox. TVB can be used in a client-server configuration, such that it can be remotely accessed through the Internet thanks to its web-based HTML5, JS, and WebGL graphical user interface. TVB is also accessible as a standalone cross-platform Python library and application, and users can interact with the scientific core through the scripting interface IDLE, enabling easy modeling, development and debugging of the scientific kernel. This second interface makes TVB extensible by combining it with other libraries and modules developed by the Python scientific community. In this article, we describe the theoretical background and foundations that led to the development of TVB, the architecture and features of its major software components as well as potential neuroscience applications.
Keywords: connectome; full-brain network model; large-scale simulation; neural masses; python; time delays; virtual brain; web platform.
Figures







References
-
- Atay F., Hutt A. (2006). Neural fields with distributed transmission speeds and long range feedback delays. SIAD 5, 670–698 10.1137/050629367 - DOI
LinkOut - more resources
Full Text Sources
Other Literature Sources

