Identification of group B respiratory syncytial viruses that lack the 60-nucleotide duplication after six consecutive epidemics of total BA dominance at coastal Kenya
- PMID: 23782406
- PMCID: PMC3963446
- DOI: 10.1111/irv.12131
Identification of group B respiratory syncytial viruses that lack the 60-nucleotide duplication after six consecutive epidemics of total BA dominance at coastal Kenya
Abstract
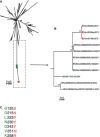
Respiratory syncytial virus BA genotype has reportedly replaced other group B genotypes worldwide. We report the observation of three group B viruses, all identical in G sequence but lacking the BA duplication, at a coastal district hospital in Kenya in early 2012. This follows a period of six consecutive respiratory syncytial virus (RSV) epidemics with 100% BA dominance among group B isolates. The new strains appear only distantly related to BA variants and to previously circulating SAB1 viruses last seen in the district in 2005, suggesting that they were circulating elsewhere undetected. These results are of relevance to an understanding of RSV persistence.
Keywords: Attachment (G gene); BA genotype; genetic diversity; respiratory syncytial virus.
© 2013 John Wiley & Sons Ltd. Influenza and Other Respiratory Viruses published by John Wiley & Sons.
Figures


References
-
- Cane P. Molecular epidemiology and evolution of RSV; in Cane P. (ed.): Respiratory Syncytial Virus. Amsterdam: Elsevier, 2007; 89–113.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

