Motion of variable-length MreB filaments at the bacterial cell membrane influences cell morphology
- PMID: 23783036
- PMCID: PMC3727927
- DOI: 10.1091/mbc.E12-10-0728
Motion of variable-length MreB filaments at the bacterial cell membrane influences cell morphology
Abstract
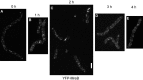
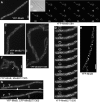
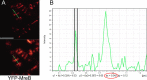
The maintenance of rod-cell shape in many bacteria depends on actin-like MreB proteins and several membrane proteins that interact with MreB. Using superresolution microscopy, we show that at 50-nm resolution, Bacillus subtilis MreB forms filamentous structures of length up to 3.4 μm underneath the cell membrane, which run at angles diverging up to 40° relative to the cell circumference. MreB from Escherichia coli forms at least 1.4-μm-long filaments. MreB filaments move along various tracks with a maximal speed of 85 nm/s, and the loss of ATPase activity leads to the formation of extended and static filaments. Suboptimal growth conditions lead to formation of patch-like structures rather than extended filaments. Coexpression of wild-type MreB with MreB mutated in the subunit interface leads to formation of shorter MreB filaments and a strong effect on cell shape, revealing a link between filament length and cell morphology. Thus MreB has an extended-filament architecture with the potential to position membrane proteins over long distances, whose localization in turn may affect the shape of the cell wall.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

