Combinatorial temporal patterning in progenitors expands neural diversity
- PMID: 23783519
- PMCID: PMC3941985
- DOI: 10.1038/nature12266
Combinatorial temporal patterning in progenitors expands neural diversity
Abstract
Human outer subventricular zone (OSVZ) neural progenitors and Drosophila type II neuroblasts both generate intermediate neural progenitors (INPs) that populate the adult cerebral cortex or central complex, respectively. It is unknown whether INPs simply expand or also diversify neural cell types. Here we show that Drosophila INPs sequentially generate distinct neural subtypes, that INPs sequentially express Dichaete, Grainy head and Eyeless transcription factors, and that these transcription factors are required for the production of distinct neural subtypes. Moreover, parental type II neuroblasts also sequentially express transcription factors and generate different neuronal/glial progeny over time, providing a second temporal identity axis. We conclude that neuroblast and INP temporal patterning axes act together to generate increased neural diversity within the adult central complex; OSVZ progenitors may use similar mechanisms to increase neural diversity in the human brain.
Figures
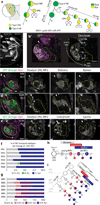

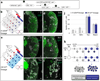
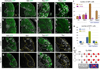
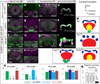
Comment in
-
Neuroscience: stem cells in multiple time zones.Nature. 2013 Jun 27;498(7455):441-3. doi: 10.1038/nature12261. Epub 2013 Jun 19. Nature. 2013. PMID: 23783512 No abstract available.
References
-
- Tanabe Y, Jessell TM. Diversity and pattern in the developing spinal cord. Science. 1996;274:1115–1123. - PubMed
-
- Krumlauf R, et al. Hox homeobox genes and regionalisation of the nervous system. Journal of neurobiology. 1993;24:1328–1340. - PubMed
-
- Maurange C, Cheng L, Gould AP. Temporal transcription factors and their targets schedule the end of neural proliferation in Drosophila. Cell. 2008;133:891–902. - PubMed
-
- Brody T, Odenwald WF. Programmed transformations in neuroblast gene expression during Drosophila CNS lineage development. Dev Biol. 2000;226:34–44. - PubMed
-
- Isshiki T, Pearson B, Holbrook S, Doe CQ. Drosophila neuroblasts sequentially express transcription factors which specify the temporal identity of their neuronal progeny. Cell. 2001;106:511–521. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

