Hepatitis C virus 3'UTR regulates viral translation through direct interactions with the host translation machinery
- PMID: 23783572
- PMCID: PMC3763534
- DOI: 10.1093/nar/gkt543
Hepatitis C virus 3'UTR regulates viral translation through direct interactions with the host translation machinery
Abstract
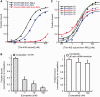
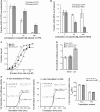
The 3' untranslated region (3'UTR) of hepatitis C virus (HCV) messenger RNA stimulates viral translation by an undetermined mechanism. We identified a high affinity interaction, conserved among different HCV genotypes, between the HCV 3'UTR and the host ribosome. The 3'UTR interacts with 40S ribosomal subunit proteins residing primarily in a localized region on the 40S solvent-accessible surface near the messenger RNA entry and exit sites. This region partially overlaps with the site where the HCV internal ribosome entry site was found to bind, with the internal ribosome entry site-40S subunit interaction being dominant. Despite its ability to bind to 40S subunits independently, the HCV 3'UTR only stimulates translation in cis, without affecting the first round translation rate. These observations support a model in which the HCV 3'UTR retains ribosome complexes during translation termination to facilitate efficient initiation of subsequent rounds of translation.
Figures




References
-
- Wells SE, Hillner PE, Vale RD, Sachs AB. Circularization of mRNA by eukaryotic translation initiation factors. Mol. Cell. 1998;2:135–140. - PubMed
-
- Komarova AV, Brocard M, Kean KM. The case for mRNA 5′ and 3′ end cross talk during translation in a eukaryotic cell. Prog. Nucleic Acid Res. Mol. Biol. 2006;81:331–367. - PubMed
-
- Gazo BM, Murphy P, Gatchel JR, Browning KS. A novel interaction of Cap-binding protein complexes eukaryotic initiation factor (eIF) 4F and eIF(iso)4F with a region in the 3′-untranslated region of satellite tobacco necrosis virus. J. Biol. Chem. 2004;279:13584–13592. - PubMed

