Protein folding and unfolding under force
- PMID: 23784721
- PMCID: PMC4065244
- DOI: 10.1002/bip.22321
Protein folding and unfolding under force
Abstract
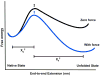
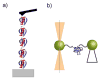
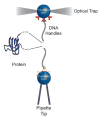
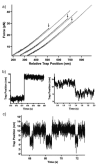
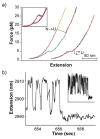
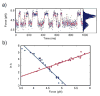
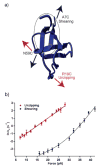
The recent revolution in optics and instrumentation has enabled the study of protein folding using extremely low mechanical forces as the denaturant. This exciting development has led to the observation of the protein folding process at single molecule resolution and its response to mechanical force. Here, we describe the principles and experimental details of force spectroscopy on proteins, with a focus on the optical tweezers instrument. Several recent results will be discussed to highlight the importance of this technique in addressing a variety of questions in the protein folding field.
Keywords: force spectroscopy; optical tweezers; protein folding.
Copyright © 2013 Wiley Periodicals, Inc.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

