p75 neurotrophin receptor is a clock gene that regulates oscillatory components of circadian and metabolic networks
- PMID: 23785138
- PMCID: PMC3685830
- DOI: 10.1523/JNEUROSCI.2757-12.2013
p75 neurotrophin receptor is a clock gene that regulates oscillatory components of circadian and metabolic networks
Abstract
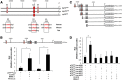
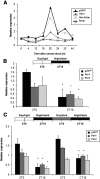
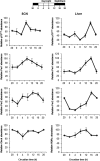
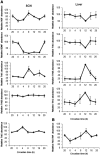
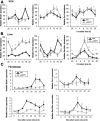
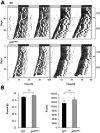
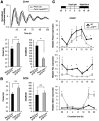
The p75 neurotrophin receptor (p75(NTR)) is a member of the tumor necrosis factor receptor superfamily with a widespread pattern of expression in tissues such as the brain, liver, lung, and muscle. The mechanisms that regulate p75(NTR) transcription in the nervous system and its expression in other tissues remain largely unknown. Here we show that p75(NTR) is an oscillating gene regulated by the helix-loop-helix transcription factors CLOCK and BMAL1. The p75(NTR) promoter contains evolutionarily conserved noncanonical E-box enhancers. Deletion mutagenesis of the p75(NTR)-luciferase reporter identified the -1039 conserved E-box necessary for the regulation of p75(NTR) by CLOCK and BMAL1. Accordingly, gel-shift assays confirmed the binding of CLOCK and BMAL1 to the p75(NTR-)1039 E-box. Studies in mice revealed that p75(NTR) transcription oscillates during dark and light cycles not only in the suprachiasmatic nucleus (SCN), but also in peripheral tissues including the liver. Oscillation of p75(NTR) is disrupted in Clock-deficient and mutant mice, is E-box dependent, and is in phase with clock genes, such as Per1 and Per2. Intriguingly, p75(NTR) is required for circadian clock oscillation, since loss of p75(NTR) alters the circadian oscillation of clock genes in the SCN, liver, and fibroblasts. Consistent with this, Per2::Luc/p75(NTR-/-) liver explants showed reduced circadian oscillation amplitude compared with those of Per2::Luc/p75(NTR+/+). Moreover, deletion of p75(NTR) also alters the circadian oscillation of glucose and lipid homeostasis genes. Overall, our findings reveal that the transcriptional activation of p75(NTR) is under circadian regulation in the nervous system and peripheral tissues, and plays an important role in the maintenance of clock and metabolic gene oscillation.
Figures



References
-
- Akhtar RA, Reddy AB, Maywood ES, Clayton JD, King VM, Smith AG, Gant TW, Hastings MH, Kyriacou CP. Circadian cycling of the mouse liver transcriptome, as revealed by cDNA microarray, is driven by the suprachiasmatic nucleus. Curr Biol. 2002;12:540–550. doi: 10.1016/S0960-9822(02)00759-5. - DOI - PubMed
-
- Baeza-Raja B, Li P, Le Moan N, Sachs BD, Schachtrup C, Davalos D, Vagena E, Bridges D, Kim C, Saltiel AR, Olefsky JM, Akassoglou K. p75 neurotrophin receptor regulates glucose homeostasis and insulin sensitivity. Proc Natl Acad Sci U S A. 2012;109:5838–5843. doi: 10.1073/pnas.1103638109. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
