Viral retasking of hBre1/RNF20 to recruit hPaf1 for transcriptional activation
- PMID: 23785282
- PMCID: PMC3681745
- DOI: 10.1371/journal.ppat.1003411
Viral retasking of hBre1/RNF20 to recruit hPaf1 for transcriptional activation
Abstract
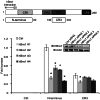
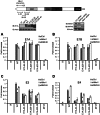
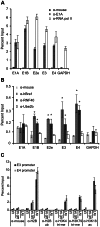
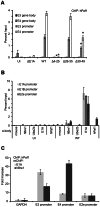
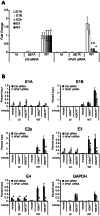
Upon infection, human adenovirus (HAdV) must activate the expression of its early genes to reprogram the cellular environment to support virus replication. This activation is orchestrated in large part by the first HAdV gene expressed during infection, early region 1A (E1A). E1A binds and appropriates components of the cellular transcriptional machinery to modulate cellular gene transcription and activate viral early genes transcription. Previously, we identified hBre1/RNF20 as a target for E1A. The interaction between E1A and hBre1 antagonizes the innate antiviral response by blocking H2B monoubiquitination, a chromatin modification necessary for the interferon (IFN) response. Here, we describe a second distinct role for the interaction of E1A with hBre1 in transcriptional activation of HAdV early genes. Furthermore, we show that E1A changes the function of hBre1 from a ubiquitin ligase involved in substrate selection to a scaffold which recruits hPaf1 as a means to stimulate transcription and transcription-coupled histone modifications. By using hBre1 to recruit hPaf1, E1A is able to optimally activate viral early transcription and begin the cycle of viral replication. The ability of E1A to target hBre1 to simultaneously repress cellular IFN dependent transcription while activating viral transcription, represents an elegant example of the incredible economy of action accomplished by a viral regulatory protein through a single protein interaction.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


References
-
- Frisch SM, Mymryk JS (2002) Adenovirus-5 E1A: paradox and paradigm. Nature Reviews Molecular Cell Biology 3: 441–452. - PubMed
-
- Hearing P, Shenk T (1986) The adenovirus type 5 E1A enhancer contains two functionally distinct domains: one is specific for E1A and the other modulates all early units in cis. Cell 45: 229–236. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

