RNA-interference components are dispensable for transcriptional silencing of the drosophila bithorax-complex
- PMID: 23785447
- PMCID: PMC3681981
- DOI: 10.1371/journal.pone.0065740
RNA-interference components are dispensable for transcriptional silencing of the drosophila bithorax-complex
Abstract
Background: Beyond their role in post-transcriptional gene silencing, Dicer and Argonaute, two components of the RNA interference (RNAi) machinery, were shown to be involved in epigenetic regulation of centromeric heterochromatin and transcriptional gene silencing. In particular, RNAi mechanisms appear to play a role in repeat induced silencing and some aspects of Polycomb-mediated gene silencing. However, the functional interplay of RNAi mechanisms and Polycomb group (PcG) pathways at endogenous loci remains to be elucidated.
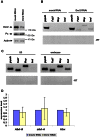
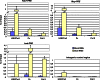
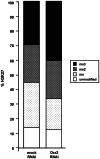
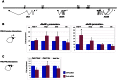
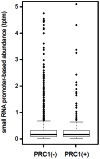
Principal findings: Here we show that the endogenous Dicer-2/Argonaute-2 RNAi pathway is dispensable for the PcG mediated silencing of the homeotic Bithorax Complex (BX-C). Although Dicer-2 depletion triggers mild transcriptional activation at Polycomb Response Elements (PREs), this does not induce transcriptional changes at PcG-repressed genes. Moreover, Dicer-2 is not needed to maintain global levels of methylation of lysine 27 of histone H3 and does not affect PRE-mediated higher order chromatin structures within the BX-C. Finally bioinformatic analysis, comparing published data sets of PcG targets with Argonaute-2-bound small RNAs reveals no enrichment of these small RNAs at promoter regions associated with PcG proteins.
Conclusions: We conclude that the Dicer-2/Argonaute-2 RNAi pathway, despite its role in pairing sensitive gene silencing of transgenes, does not have a role in PcG dependent silencing of major homeotic gene cluster loci in Drosophila.
Conflict of interest statement
Figures

References
-
- Bartel DP (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116: 281–297. - PubMed
-
- Baulcombe D (2004) RNA silencing in plants. Nature 431: 356–363. - PubMed
-
- Meister G, Tuschl T (2004) Mechanisms of gene silencing by double-stranded RNA. Nature 431: 343–349. - PubMed
-
- Lee YS, Nakahara K, Pham JW, Kim K, He Z, et al. (2004) Distinct roles for Drosophila Dicer-1 and Dicer-2 in the siRNA/miRNA silencing pathways. Cell 117: 69–81. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

