Transcription poisoning by Topoisomerase I is controlled by gene length, splice sites, and miR-142-3p
- PMID: 23786772
- PMCID: PMC3874869
- DOI: 10.1158/0008-5472.CAN-12-3504
Transcription poisoning by Topoisomerase I is controlled by gene length, splice sites, and miR-142-3p
Abstract
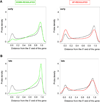
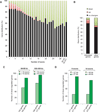
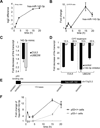
Topoisomerase I (Top1) relaxes DNA supercoiling by forming transient cleavage complexes (Top1cc) up- and downstream of transcription complexes. Top1cc can be trapped by carcinogenic and endogenous DNA lesions and by camptothecin, resulting in transcription blocks. Here, we undertook genome-wide analysis of camptothecin-treated cells at exon resolution. RNA samples from HCT116 and MCF7 cells were analyzed with the Affy Exon Array platform, allowing high-resolution mapping along 18,537 genes. Long genes that are highly expressed were the most susceptible to downregulation, whereas short genes were preferentially upregulated. Along the body of genes, downregulation was most important toward the 3'-end and increased with the number of exon-intron junctions. Ubiquitin and RNA degradation-related pathway genes were selectively downregulated. Parallel analysis of microRNA with the Agilent miRNA microarray platform revealed that miR-142-3p was highly induced by camptothecin. More than 10% of the downregulated genes were targets of this p53-dependent microRNA. Our study shows the profound impact of Top1cc on transcription elongation, especially at intron-exon junctions and on transcript stability by microRNA miR-142-3p upregulation.
©2013 AACR.
Conflict of interest statement
Figures



References
-
- Pommier Y. Topoisomerase I inhibitors: camptothecins and beyond. Nat Rev Cancer. 2006;6:789–802. - PubMed
-
- Hsiang YH, Hertzberg R, Hecht S, Liu LF. Camptothecin induces protein-linked DNA breaks via mammalian DNA topoisomerase I. J Biol Chem. 1985;260:14873–14878. - PubMed
-
- Ljungman M, Hanawalt PC. The anti-cancer drug camptothecin inhibits elongation but stimulates initiation of RNA polymerase II transcription. Carcinogenesis. 1996;17:31–35. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

