Hydrocarbon-degrading bacteria enriched by the Deepwater Horizon oil spill identified by cultivation and DNA-SIP
- PMID: 23788333
- PMCID: PMC3806270
- DOI: 10.1038/ismej.2013.98
Hydrocarbon-degrading bacteria enriched by the Deepwater Horizon oil spill identified by cultivation and DNA-SIP
Abstract
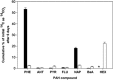
The massive influx of crude oil into the Gulf of Mexico during the Deepwater Horizon (DWH) disaster triggered dramatic microbial community shifts in surface oil slick and deep plume waters. Previous work had shown several taxa, notably DWH Oceanospirillales, Cycloclasticus and Colwellia, were found to be enriched in these waters based on their dominance in conventional clone and pyrosequencing libraries and were thought to have had a significant role in the degradation of the oil. However, this type of community analysis data failed to provide direct evidence on the functional properties, such as hydrocarbon degradation of organisms. Using DNA-based stable-isotope probing with uniformly (13)C-labelled hydrocarbons, we identified several aliphatic (Alcanivorax, Marinobacter)- and polycyclic aromatic hydrocarbon (Alteromonas, Cycloclasticus, Colwellia)-degrading bacteria. We also isolated several strains (Alcanivorax, Alteromonas, Cycloclasticus, Halomonas, Marinobacter and Pseudoalteromonas) with demonstrable hydrocarbon-degrading qualities from surface slick and plume water samples collected during the active phase of the spill. Some of these organisms accounted for the majority of sequence reads representing their respective taxa in a pyrosequencing data set constructed from the same and additional water column samples. Hitherto, Alcanivorax was not identified in any of the previous water column studies analysing the microbial response to the spill and we discuss its failure to respond to the oil. Collectively, our data provide unequivocal evidence on the hydrocarbon-degrading qualities for some of the dominant taxa enriched in surface and plume waters during the DWH oil spill, and a more complete understanding of their role in the fate of the oil.
Figures


References
-
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. - PubMed
-
- Bælum J, Borglin S, Chakraborty R, Fortney JL, Lamendella R, Mason OU, et al. Deep-sea bacteria enriched by oil and dispersant from the Deepwater Horizon Spill. Environ Microbiol. 2012;14:2405–2416. - PubMed
-
- Camilli R, Reddy CM, Yoerger DR, Van Mooy BAS, Jakuba MV, Kinsey JC, et al. Tracking hydrocarbon plume transport and biodegradation at Deepwater Horizon. Science. 2010;330:201–204. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

