Isolation of αL I domain mutants mediating firm cell adhesion using a novel flow-based sorting method
- PMID: 23788664
- PMCID: PMC3711393
- DOI: 10.1093/protein/gzt028
Isolation of αL I domain mutants mediating firm cell adhesion using a novel flow-based sorting method
Abstract
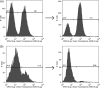
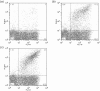
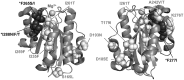
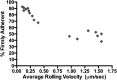
The inserted (I) domain of αLβ2 integrin (LFA-1) contains the entire binding site of the molecule. It mediates both rolling and firm adhesion of leukocytes at sites of inflammation depending on the activation state of the integrin. The affinity change of the entire integrin can be mimicked by the I domain alone through mutations that affect the conformation of the molecule. High-affinity mutants of the I domain have been discovered previously using both rational design and directed evolution. We have found that binding affinity fails to dictate the behavior of I domain adhesion under shear flow. In order to better understand I domain adhesion, we have developed a novel panning method to separate yeast expressing a library of I domain variants on the surface by adhesion under flow. Using conditions analogous to those experienced by cells interacting with the post-capillary vascular endothelium, we have identified mutations supporting firm adhesion that are not found using typical directed evolution techniques that select for tight binding to soluble ligands. Mutants isolated using this method do not cluster with those found by sorting with soluble ligand. Furthermore, these mutants mediate shear-driven cell rolling dynamics decorrelated from binding affinity, as previously observed for I domains bearing engineered disulfide bridges to stabilize activated conformational states. Characterization of these mutants supports a greater understanding of the structure-function relationship of the αL I domain, and of the relationship between applied force and bioadhesion in a broader context.
Keywords: I domain; conformational regulation; rolling adhesion; yeast display; αL integrin.
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

