A novel insight into the mechanism of mammalian selenoprotein synthesis
- PMID: 23788723
- PMCID: PMC3708534
- DOI: 10.1261/rna.036871.112
A novel insight into the mechanism of mammalian selenoprotein synthesis
Abstract
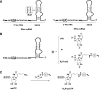
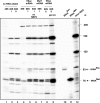
The amino acid selenocysteine is encoded by UGA, usually a stop codon, thus requiring a specialized machinery to enable its incorporation into selenoproteins. The machinery comprises the tRNA(Sec), a 3'-UTR mRNA stem-loop termed SElenoCysteine Insertion Sequence (SECIS), which is mandatory for recoding UGA as a Sec codon, the SECIS Binding Protein 2 (SBP2), and other proteins. Little is known about the molecular mechanism and, in particular, when, where, and how the SECIS and SBP2 contact the ribosome. Previous work by others used the isolated SECIS RNA to address this question. Here, we developed a novel approach using instead engineered minimal selenoprotein mRNAs containing SECIS elements derivatized with photoreactive groups. By cross-linking experiments in rabbit reticulocyte lysate, new information could be gained about the SBP2 and SECIS contacts with components of the translation machinery at various translation steps. In particular, we found that SBP2 was bound only to the SECIS in 48S pre-initiation and 80S pretranslocation complexes. In the complex where the Sec-tRNA(Sec) was accommodated to the A site but transpeptidation was blocked, SBP2 bound the ribosome and possibly the SECIS element as well, and the SECIS had flexible contacts with the 60S ribosomal subunit involving several ribosomal proteins. Altogether, our findings led to broadening our understanding about the unique mechanism of selenocysteine incorporation in mammals.
Keywords: SECIS-binding protein 2; cross-linking approach; mammalian ribosome; selenocysteine incorporation; selenocysteine insertion sequence.
Figures




References
-
- Allmang C, Wurth L, Krol A 2009. The selenium to selenoproteins pathway in eukaryotes: More molecular pathways than anticipated. Biochim Biophys Acta 1790: 1415–1423 - PubMed
-
- Anthony DD, Merrick WC 1992. Analysis of 40S and 80S complexes with mRNA as measured by sucrose density gradients and primer extension inhibition. J Biol Chem 267: 1554–1562 - PubMed
-
- Böck A, Rother M, Leibundgut M, Ban N 2006. Selenium metabolism in prokaryotes. In Selenium: Its molecular biology and role in human health, 2nd ed. (ed. Hatfield DL, et al.), pp. 9–28 Springer Verlag, New York
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
