Jpx RNA activates Xist by evicting CTCF
- PMID: 23791181
- PMCID: PMC3777401
- DOI: 10.1016/j.cell.2013.05.028
Jpx RNA activates Xist by evicting CTCF
Abstract
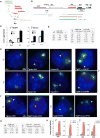
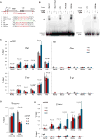
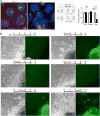
In mammals, dosage compensation between XX and XY individuals occurs through X chromosome inactivation (XCI). The noncoding Xist RNA is expressed and initiates XCI only when more than one X chromosome is present. Current models invoke a dependency on the X-to-autosome ratio (X:A), but molecular factors remain poorly defined. Here, we demonstrate that molecular titration between an X-encoded RNA and an autosomally encoded protein dictates Xist induction. In pre-XCI cells, CTCF protein represses Xist transcription. At the onset of XCI, Jpx RNA is upregulated, binds CTCF, and extricates CTCF from one Xist allele. We demonstrate that CTCF is an RNA-binding protein and is titrated away from the Xist promoter by Jpx RNA. Thus, Jpx activates Xist by evicting CTCF. The functional antagonism via molecular titration reveals a role for long noncoding RNA in epigenetic regulation.
Copyright © 2013 Elsevier Inc. All rights reserved.
Figures




Comment in
-
Epigenetics: X inactivation by titration.Nat Rev Genet. 2013 Aug;14(8):518. doi: 10.1038/nrg3538. Epub 2013 Jul 2. Nat Rev Genet. 2013. PMID: 23817312 No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

