Polarity-dependent distribution of angiomotin localizes Hippo signaling in preimplantation embryos
- PMID: 23791731
- PMCID: PMC3742369
- DOI: 10.1016/j.cub.2013.05.014
Polarity-dependent distribution of angiomotin localizes Hippo signaling in preimplantation embryos
Abstract
Background: In preimplantation mouse embryos, the first cell fate specification to the trophectoderm or inner cell mass occurs by the early blastocyst stage. The cell fate is controlled by cell position-dependent Hippo signaling, although the mechanisms underlying position-dependent Hippo signaling are unknown.
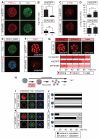
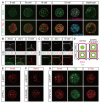
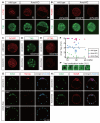
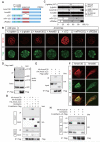
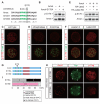
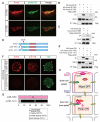
Results: We show that a combination of cell polarity and cell-cell adhesion establishes position-dependent Hippo signaling, where the outer and inner cells are polar and nonpolar, respectively. The junction-associated proteins angiomotin (Amot) and angiomotin-like 2 (Amotl2) are essential for Hippo pathway activation and appropriate cell fate specification. In the nonpolar inner cells, Amot localizes to adherens junctions (AJs), and cell-cell adhesion activates the Hippo pathway. In the outer cells, the cell polarity sequesters Amot from basolateral AJs to apical domains, thereby suppressing Hippo signaling. The N-terminal domain of Amot is required for actin binding, Nf2/Merlin-mediated association with the E-cadherin complex, and interaction with Lats protein kinase. In AJs, S176 in the N-terminal domain of Amot is phosphorylated by Lats, which inhibits the actin-binding activity, thereby stabilizing the Amot-Lats interaction to activate the Hippo pathway.
Conclusions: We propose that the phosphorylation of S176 in Amot is a critical step for activation of the Hippo pathway in AJs and that cell polarity disconnects the Hippo pathway from cell-cell adhesion by sequestering Amot from AJs. This mechanism converts positional information into differential Hippo signaling, thereby leading to differential cell fates.
Copyright © 2013 Elsevier Ltd. All rights reserved.
Figures
Comment in
-
Development: Hippo signalling turns the embryo inside out.Curr Biol. 2013 Jul 8;23(13):R559-61. doi: 10.1016/j.cub.2013.05.064. Curr Biol. 2013. PMID: 23845241
References
-
- Tarkowski AK, Wroblewska J. Development of blastomeres of mouse eggs isolated at the 4- and 8-cell stage. Journal of embryology and experimental morphology. 1967;18:155–180. - PubMed
-
- Johnson MH, Ziomek CA. The foundation of two distinct cell lineages within the mouse morula. Cell. 1981;24:71–80. - PubMed
-
- Yamanaka Y, Ralston A, Stephenson RO, Rossant J. Cell and molecular regulation of the mouse blastocyst. Dev Dyn. 2006;235:2301–2314. - PubMed
-
- Niwa H, Toyooka Y, Shimosato D, Strumpf D, Takahashi K, Yagi R, Rossant J. Interaction between Oct3/4 and Cdx2 determines trophectoderm differentiation. Cell. 2005;123:917–929. - PubMed
-
- Nishioka N, Yamamoto S, Kiyonari H, Sato H, Sawada A, Ota M, Nakao K, Sasaki H. Tead4 is required for specification of trophectoderm in pre-implantation mouse embryos. Mechanisms of development. 2008;125:270–283. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

