Promoter directionality is controlled by U1 snRNP and polyadenylation signals
- PMID: 23792564
- PMCID: PMC3720719
- DOI: 10.1038/nature12349
Promoter directionality is controlled by U1 snRNP and polyadenylation signals
Abstract
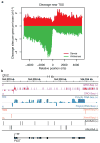
Transcription of the mammalian genome is pervasive, but productive transcription outside of protein-coding genes is limited by unknown mechanisms. In particular, although RNA polymerase II (RNAPII) initiates divergently from most active gene promoters, productive elongation occurs primarily in the sense-coding direction. Here we show in mouse embryonic stem cells that asymmetric sequence determinants flanking gene transcription start sites control promoter directionality by regulating promoter-proximal cleavage and polyadenylation. We find that upstream antisense RNAs are cleaved and polyadenylated at poly(A) sites (PASs) shortly after initiation. De novo motif analysis shows PAS signals and U1 small nuclear ribonucleoprotein (snRNP) recognition sites to be the most depleted and enriched sequences, respectively, in the sense direction relative to the upstream antisense direction. These U1 snRNP sites and PAS sites are progressively gained and lost, respectively, at the 5' end of coding genes during vertebrate evolution. Functional disruption of U1 snRNP activity results in a dramatic increase in promoter-proximal cleavage events in the sense direction with slight increases in the antisense direction. These data suggest that a U1-PAS axis characterized by low U1 snRNP recognition and a high density of PASs in the upstream antisense region reinforces promoter directionality by promoting early termination in upstream antisense regions, whereas proximal sense PAS signals are suppressed by U1 snRNP. We propose that the U1-PAS axis limits pervasive transcription throughout the genome.
Conflict of interest statement
Figures



References
-
- Preker P, et al. RNA exosome depletion reveals transcription upstream of active human promoters. Science. 2008;322:1851–1854. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

