Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform
- PMID: 23793624
- PMCID: PMC3753973
- DOI: 10.1128/AEM.01043-13
Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform
Abstract
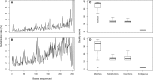
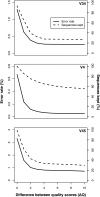
Rapid advances in sequencing technology have changed the experimental landscape of microbial ecology. In the last 10 years, the field has moved from sequencing hundreds of 16S rRNA gene fragments per study using clone libraries to the sequencing of millions of fragments per study using next-generation sequencing technologies from 454 and Illumina. As these technologies advance, it is critical to assess the strengths, weaknesses, and overall suitability of these platforms for the interrogation of microbial communities. Here, we present an improved method for sequencing variable regions within the 16S rRNA gene using Illumina's MiSeq platform, which is currently capable of producing paired 250-nucleotide reads. We evaluated three overlapping regions of the 16S rRNA gene that vary in length (i.e., V34, V4, and V45) by resequencing a mock community and natural samples from human feces, mouse feces, and soil. By titrating the concentration of 16S rRNA gene amplicons applied to the flow cell and using a quality score-based approach to correct discrepancies between reads used to construct contigs, we were able to reduce error rates by as much as two orders of magnitude. Finally, we reprocessed samples from a previous study to demonstrate that large numbers of samples could be multiplexed and sequenced in parallel with shotgun metagenomes. These analyses demonstrate that our approach can provide data that are at least as good as that generated by the 454 platform while providing considerably higher sequencing coverage for a fraction of the cost.
Figures


References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

