The Crohn's disease: associated ATG16L1 variant and Salmonella invasion
- PMID: 23794574
- PMCID: PMC3686164
- DOI: 10.1136/bmjopen-2013-002790
The Crohn's disease: associated ATG16L1 variant and Salmonella invasion
Erratum in
-
Correction.BMJ Open. 2015 Jul 27;5(7):e002790corr1. doi: 10.1136/bmjopen-2013-002790corr1. BMJ Open. 2015. PMID: 26216149 Free PMC article. No abstract available.
Abstract
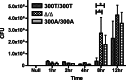
Objective: A common genetic coding variant in the core autophagy gene ATG16L1 is associated with increased susceptibility to Crohn's disease (CD). The variant encodes an amino acid change in ATG16L1 such that the threonine at position 300 is substituted with an alanine (ATG16L1 T300A). How this variant contributes to increased risk of CD is not known, but studies with transfected cell lines and gene-targeted mice have demonstrated that ATG16L1 is required for autophagy, control of interleukin-1-β and autophagic clearance of intracellular microbes. In addition, studies with human cells expressing ATG16L1 T300A indicate that this variant reduces the autophagic clearance of intracellular microbes.
Design/results: We demonstrate, using somatically gene-targeted human cells that the ATG16L1 T300A variant confers protection from cellular invasion by Salmonella. In addition, we show that ATG16L1-deficient cells are resistant to bacterial invasion.
Conclusions: These results suggest that cellular expression of ATG16L1 facilitates bacterial invasion and that the CD-associated ATG16L1 T300A variant may confer protection from bacterial infection.
Figures


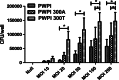
References
-
- Hampe J, Franke A, Rosenstiel P, et al. A genome-wide association scan of nonsynonymous SNPs identifies a susceptibility variant for Crohn disease in ATG16L1. Nat Genet 2007;39:207–11 - PubMed
-
- Ohsumi Y. Molecular dissection of autophagy: two ubiquitin-like systems. Nat Rev Mol Cell Biol 2001;2:211–16 - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
