The homologous recombination machinery modulates the formation of RNA-DNA hybrids and associated chromosome instability
- PMID: 23795288
- PMCID: PMC3679537
- DOI: 10.7554/eLife.00505
The homologous recombination machinery modulates the formation of RNA-DNA hybrids and associated chromosome instability
Abstract
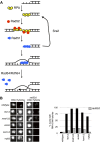
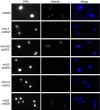
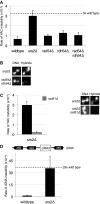
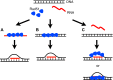
Genome instability in yeast and mammals is caused by RNA-DNA hybrids that form as a result of defects in different aspects of RNA biogenesis. We report that in yeast mutants defective for transcription repression and RNA degradation, hybrid formation requires Rad51p and Rad52p. These proteins normally promote DNA-DNA strand exchange in homologous recombination. We suggest they also directly promote the DNA-RNA strand exchange necessary for hybrid formation since we observed accumulation of Rad51p at a model hybrid-forming locus. Furthermore, we provide evidence that Rad51p mediates hybridization of transcripts to homologous chromosomal loci distinct from their site of synthesis. This hybrid formation in trans amplifies the genome-destabilizing potential of RNA and broadens the exclusive co-transcriptional models that pervade the field. The deleterious hybrid-forming activity of Rad51p is counteracted by Srs2p, a known Rad51p antagonist. Thus Srs2p serves as a novel anti-hybrid mechanism in vivo. DOI:http://dx.doi.org/10.7554/eLife.00505.001.
Keywords: DNA repair; R loops; RNA-DNA hybrids; Rad51; S. cerevisiae; genome instability.
Conflict of interest statement
The authors declare that no competing interests exist.
Figures






Comment in
-
Rad51, friend or foe?Elife. 2013 Jun 11;2:e00914. doi: 10.7554/eLife.00914. Elife. 2013. PMID: 23795299 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

