Skin microbiome imbalance in patients with STAT1/STAT3 defects impairs innate host defense responses
- PMID: 23796786
- PMCID: PMC4045018
- DOI: 10.1159/000351912
Skin microbiome imbalance in patients with STAT1/STAT3 defects impairs innate host defense responses
Abstract
Background: Chronic mucocutaneous candidiasis (CMC) and hyper-IgE syndrome (HIES) are primary immunodeficiencies mainly caused by mutations in STAT1 and STAT3, respectively. CMC and HIES patients have an increased risk for skin and mucosal infections with fungal pathogens and Staphylococcus aureus. However, it is unknown whether the genetic defects in these patients also affect the skin and mucosal microbiome, which in turn may influence host defense mechanisms.
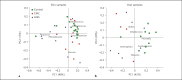
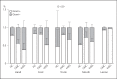
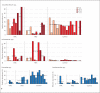
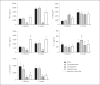
Methods: The skin and oral microbiome of CMC and HIES patients was compared to that of healthy controls at five body sites using 16S rRNA sequencing. The influence of skin colonizers on the immune response was investigated using in vitro experiments.
Results: The microbiome of CMC and HIES patients contained more Gram-negative bacteria, especially Acinetobacter spp., and less of the normal Corynebacterium spp. compared to healthy controls. Exposure of human primary leukocytes to Acinetobacter suppressed the cytokine response to Candida albicans and S. aureus, while the normal corynebacteria did not suppress cytokine responses.
Discussion: These results demonstrate that central mediators of immune responses like STAT1 and STAT3 not only directly influence immune responses, but also result in changes in the skin microbiome that in turn can amplify the defective immune response against fungal and microbial pathogens.
Copyright © 2013 S. Karger AG, Basel.
Figures
References
-
- Kirkpatrick CH. Chronic mucocutaneous candidiasis. J Am Acad Dermatol. 1994;31:S14–S17. - PubMed
-
- Aaltonen J, Björses P, Perheentupa J, Horelli Kuitunen N, Palotie A, Peltonen L, et al. An autoimmune disease, APECED, caused by mutations in a novel gene featuring two PHD-type zinc-finger domains. Nat Genet. 1997;17:399–403. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

