A survey of intragenic breakpoints in glioblastoma identifies a distinct subset associated with poor survival
- PMID: 23796897
- PMCID: PMC3713427
- DOI: 10.1101/gad.213686.113
A survey of intragenic breakpoints in glioblastoma identifies a distinct subset associated with poor survival
Abstract
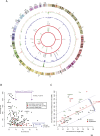
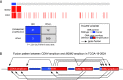
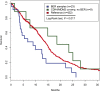
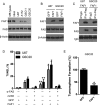
With the advent of high-throughput sequencing technologies, much progress has been made in the identification of somatic structural rearrangements in cancer genomes. However, characterization of the complex alterations and their associated mechanisms remains inadequate. Here, we report a comprehensive analysis of whole-genome sequencing and DNA copy number data sets from The Cancer Genome Atlas to relate chromosomal alterations to imbalances in DNA dosage and describe the landscape of intragenic breakpoints in glioblastoma multiforme (GBM). Gene length, guanine-cytosine (GC) content, and local presence of a copy number alteration were closely associated with breakpoint susceptibility. A dense pattern of repeated focal amplifications involving the murine double minute 2 (MDM2)/cyclin-dependent kinase 4 (CDK4) oncogenes and associated with poor survival was identified in 5% of GBMs. Gene fusions and rearrangements were detected concomitant within the breakpoint-enriched region. At the gene level, we noted recurrent breakpoints in genes such as apoptosis regulator FAF1. Structural alterations of the FAF1 gene disrupted expression and led to protein depletion. Restoration of the FAF1 protein in glioma cell lines significantly increased the FAS-mediated apoptosis response. Our study uncovered a previously underappreciated genomic mechanism of gene deregulation that can confer growth advantages on tumor cells and may generate cancer-specific vulnerabilities in subsets of GBM.
Keywords: Glioblastoma multiforme; The Cancer Genome Atlas; gene fusion; genomic instability; genomic rearrangement; intragenic breakpoint.
Figures

Comment in
-
Intragenic breakpoint: a marker of genome instability in glioblastoma.Cell Cycle. 2013 Dec 15;12(24):3705-6. doi: 10.4161/cc.26874. Epub 2013 Oct 21. Cell Cycle. 2013. PMID: 24145229 Free PMC article. No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous
