miR-9 is an essential oncogenic microRNA specifically overexpressed in mixed lineage leukemia-rearranged leukemia
- PMID: 23798388
- PMCID: PMC3710804
- DOI: 10.1073/pnas.1310144110
miR-9 is an essential oncogenic microRNA specifically overexpressed in mixed lineage leukemia-rearranged leukemia
Abstract
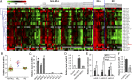
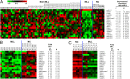
MicroRNAs (miRNAs), small noncoding RNAs that regulate target gene mRNAs, are known to contribute to pathogenesis of cancers. Acute myeloid leukemia (AML) is a group of heterogeneous hematopoietic malignancies with various chromosomal and/or molecular abnormalities. AML with chromosomal translocations involving the mixed lineage leukemia (MLL) gene are usually associated with poor survival. In the present study, through a large-scale, genomewide miRNA expression assay, we show that microRNA-9 (miR-9) is the most specifically up-regulated miRNA in MLL-rearranged AML compared with both normal control and non-MLL-rearranged AML. We demonstrate that miR-9 is a direct target of MLL fusion proteins and can be significantly up-regulated in expression by the latter in human and mouse hematopoietic stem/progenitor cells. Depletion of endogenous miR-9 expression by an appropriate antagomiR can significantly inhibit cell growth/viability and promote apoptosis in human MLL-rearranged AML cells, and the opposite is true when expression of miR-9 is forced. Blocking endogenous miR-9 function by anti-miRNA sponge can significantly inhibit, whereas forced expression of miR-9 can significantly promote, MLL fusion-induced immortalization/transformation of normal mouse bone marrow progenitor cells in vitro. Furthermore, forced expression of miR-9 can significantly promote MLL fusion-mediated leukemogenesis in vivo. In addition, a group of putative target genes of miR-9 exhibited a significant inverse correlation of expression with miR-9 in a series of leukemia sample sets, suggesting that they are potential targets of miR-9 in MLL-rearranged AML. Collectively, our data demonstrate that miR-9 is a critical oncomiR in MLL-rearranged AML and can serve as a potential therapeutic target to treat this dismal disease.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



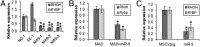
References
-
- He L, Hannon GJ. MicroRNAs: Small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5(7):522–531. - PubMed
-
- Xiao C, Rajewsky K. MicroRNA control in the immune system: Basic principles. Cell. 2009;136(1):26–36. - PubMed
-
- Löwenberg B, Downing JR, Burnett A. Acute myeloid leukemia. N Engl J Med. 1999;341(14):1051–1062. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

