Quantitative dissection of hydrogen bond-mediated proton transfer in the ketosteroid isomerase active site
- PMID: 23798390
- PMCID: PMC3710806
- DOI: 10.1073/pnas.1302191110
Quantitative dissection of hydrogen bond-mediated proton transfer in the ketosteroid isomerase active site
Abstract
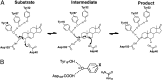
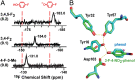
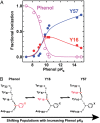
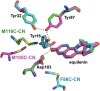
Hydrogen bond networks are key elements of protein structure and function but have been challenging to study within the complex protein environment. We have carried out in-depth interrogations of the proton transfer equilibrium within a hydrogen bond network formed to bound phenols in the active site of ketosteroid isomerase. We systematically varied the proton affinity of the phenol using differing electron-withdrawing substituents and incorporated site-specific NMR and IR probes to quantitatively map the proton and charge rearrangements within the network that accompany incremental increases in phenol proton affinity. The observed ionization changes were accurately described by a simple equilibrium proton transfer model that strongly suggests the intrinsic proton affinity of one of the Tyr residues in the network, Tyr16, does not remain constant but rather systematically increases due to weakening of the phenol-Tyr16 anion hydrogen bond with increasing phenol proton affinity. Using vibrational Stark spectroscopy, we quantified the electrostatic field changes within the surrounding active site that accompany these rearrangements within the network. We were able to model these changes accurately using continuum electrostatic calculations, suggesting a high degree of conformational restriction within the protein matrix. Our study affords direct insight into the physical and energetic properties of a hydrogen bond network within a protein interior and provides an example of a highly controlled system with minimal conformational rearrangements in which the observed physical changes can be accurately modeled by theoretical calculations.
Keywords: active site environment; computational modeling; enzyme catalysis; protein electrostatics; protein semisynthesis.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

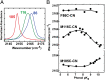
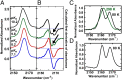

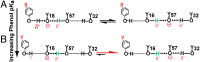
References
-
- Fersht AR. Structure and Mechanism in Protein Science. New York: Freeman; 1999.
-
- Ladd MFC, Palmer RA. Structure Determination by X-Ray Crystallography. New York: Kluwer Academic/Plenum; 2003.
-
- Antosiewicz J, McCammon JA, Gilson MK. The determinants of pKas in proteins. Biochemistry. 1996;35(24):7819–7833. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

