Molecular basis for chromatin binding and regulation of MLL5
- PMID: 23798402
- PMCID: PMC3710826
- DOI: 10.1073/pnas.1310156110
Molecular basis for chromatin binding and regulation of MLL5
Abstract
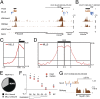
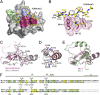
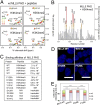
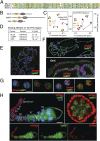
The human mixed-lineage leukemia 5 (MLL5) protein mediates hematopoietic cell homeostasis, cell cycle, and survival; however, the molecular basis underlying MLL5 activities remains unknown. Here, we show that MLL5 is recruited to gene-rich euchromatic regions via the interaction of its plant homeodomain finger with the histone mark H3K4me3. The 1.48-Å resolution crystal structure of MLL5 plant homeodomain in complex with the H3K4me3 peptide reveals a noncanonical binding mechanism, whereby K4me3 is recognized through a single aromatic residue and an aspartate. The binding induces a unique His-Asp swapping rearrangement mediated by a C-terminal α-helix. Phosphorylation of H3T3 and H3T6 abrogates the association with H3K4me3 in vitro and in vivo, releasing MLL5 from chromatin in mitosis. This regulatory switch is conserved in the Drosophila ortholog of MLL5, UpSET, and suggests the developmental control for targeting of H3K4me3. Together, our findings provide first insights into the molecular basis for the recruitment, exclusion, and regulation of MLL5 at chromatin.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Emerling BM, et al. MLL5, a homolog of Drosophila trithorax located within a segment of chromosome band 7q22 implicated in myeloid leukemia. Oncogene. 2002;21(31):4849–4854. - PubMed
-
- Heuser M, et al. Loss of MLL5 results in pleiotropic hematopoietic defects, reduced neutrophil immune function, and extreme sensitivity to DNA demethylation. Blood. 2009;113(7):1432–1443. - PubMed
-
- Madan V, et al. Impaired function of primitive hematopoietic cells in mice lacking the mixed-lineage-leukemia homolog MLL5. Blood. 2009;113(7):1444–1454. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
- R01 GM068088/GM/NIGMS NIH HHS/United States
- T32CA09156/CA/NCI NIH HHS/United States
- P30 CA046934/CA/NCI NIH HHS/United States
- R01 HL065440/HL/NHLBI NIH HHS/United States
- P30 DK056465/DK/NIDDK NIH HHS/United States
- HL65440/HL/NHLBI NIH HHS/United States
- GM068088/GM/NIGMS NIH HHS/United States
- R01 GM096863/GM/NIGMS NIH HHS/United States
- GM097083/GM/NIGMS NIH HHS/United States
- R01 GM101664/GM/NIGMS NIH HHS/United States
- GM096863/GM/NIGMS NIH HHS/United States
- T32 CA009156/CA/NCI NIH HHS/United States
- R01 GM097083/GM/NIGMS NIH HHS/United States
- GM101664/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

