Probing the transient dark state of substrate binding to GroEL by relaxation-based solution NMR
- PMID: 23798407
- PMCID: PMC3710837
- DOI: 10.1073/pnas.1305715110
Probing the transient dark state of substrate binding to GroEL by relaxation-based solution NMR
Abstract
The mechanism whereby the prototypical chaperonin GroEL performs work on substrate proteins has not yet been fully elucidated, hindered by lack of detailed structural and dynamic information on the bound substrate. Previous investigations have produced conflicting reports on the state of GroEL-bound polypeptides, largely due to the transient and dynamic nature of these complexes. Here, we present a unique approach, based on combined analysis of four complementary relaxation-based NMR experiments, to probe directly the "dark" NMR-invisible state of the model, intrinsically disordered, polypeptide amyloid β (Aβ40) bound to GroEL. The four NMR experiments, lifetime line-broadening, dark-state exchange saturation transfer, relaxation dispersion, and small exchange-induced chemical shifts, are dependent in different ways on the overall exchange rates and populations of the free and bound states of the substrate, as well as on residue-specific dynamics and structure within the bound state as reported by transverse magnetization relaxation rates and backbone chemical shifts, respectively. Global fitting of all the NMR data shows that the complex is transient with a lifetime of <1 ms, that binding involves two predominantly hydrophobic segments corresponding to predicted GroEL consensus binding sequences, and that the structure of the bound polypeptide remains intrinsically and dynamically disordered with minimal changes in secondary structure propensity relative to the free state. Our results establish a unique method to observe NMR-invisible dynamic states of GroEL-bound substrates and to describe at atomic resolution the events between substrate binding and encapsulation that are crucial for understanding the normal and stress-related metabolic function of chaperonins.
Keywords: conformational sampling; protein–protein interactions; supramolecular machine.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
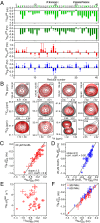
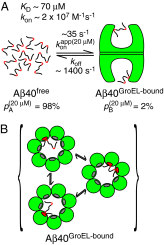
 and the populations of free (pA) and GroEL-bound (pB) Aβ40 are those obtained in the presence of 20 μM GroEL. The equilibrium dissociation constant Kd and the second-order association rate constant kon are calculated assuming each GroEL cavity only accommodates a single molecule of Aβ40 with numerous available binding modes. (B) Rapid interconversion (with a rate constant >koff) between different GroEL-bound configurations of Aβ40 consisting of the central hydrophobic, C-terminal hydrophobic, or both hydrophobic regions in contact with GroEL is possible.
and the populations of free (pA) and GroEL-bound (pB) Aβ40 are those obtained in the presence of 20 μM GroEL. The equilibrium dissociation constant Kd and the second-order association rate constant kon are calculated assuming each GroEL cavity only accommodates a single molecule of Aβ40 with numerous available binding modes. (B) Rapid interconversion (with a rate constant >koff) between different GroEL-bound configurations of Aβ40 consisting of the central hydrophobic, C-terminal hydrophobic, or both hydrophobic regions in contact with GroEL is possible.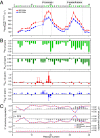
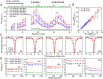
 , for GroEL-bound Aβ40 at spectrometer frequencies of 600 (blue) and 900 (red) MHz. (B) Residue-specific 15N, 1HN, 13Cα, and 13Cβ chemical shift differences between GroEL-bound and free Aβ40. The 15N chemical shift differences (15N-Δδ) are optimized in the fitting procedure, whereas the other chemical shift differences are calculated from the ΔR2 values and fitted global kinetic parameters (
, for GroEL-bound Aβ40 at spectrometer frequencies of 600 (blue) and 900 (red) MHz. (B) Residue-specific 15N, 1HN, 13Cα, and 13Cβ chemical shift differences between GroEL-bound and free Aβ40. The 15N chemical shift differences (15N-Δδ) are optimized in the fitting procedure, whereas the other chemical shift differences are calculated from the ΔR2 values and fitted global kinetic parameters (References
-
- Hendrick JP, Hartl FU. Molecular chaperone functions of heat-shock proteins. Annu Rev Biochem. 1993;62:349–384. - PubMed
-
- Thirumalai D, Lorimer GH. Chaperonin-mediated protein folding. Annu Rev Biophys Biomol Struct. 2001;30:245–269. - PubMed
-
- Horwich AL, Fenton WA. Chaperonin-mediated protein folding: Using a central cavity to kinetically assist polypeptide chain folding. Q Rev Biophys. 2009;42(2):83–116. - PubMed
-
- Saibil HR. Chaperone machines in action. Curr Opin Struct Biol. 2008;18(1):35–42. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

