Structural and functional insights into caseinolytic proteases reveal an unprecedented regulation principle of their catalytic triad
- PMID: 23798410
- PMCID: PMC3710848
- DOI: 10.1073/pnas.1219125110
Structural and functional insights into caseinolytic proteases reveal an unprecedented regulation principle of their catalytic triad
Abstract
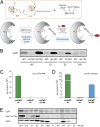
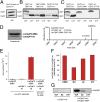
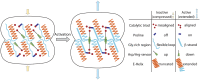
Caseinolytic proteases (ClpPs) are large oligomeric protein complexes that contribute to cell homeostasis as well as virulence regulation in bacteria. Although most organisms possess a single ClpP protein, some organisms encode two or more ClpP isoforms. Here, we elucidated the crystal structures of ClpP1 and ClpP2 from pathogenic Listeria monocytogenes and observe an unprecedented regulation principle by the catalytic triad. Whereas L. monocytogenes (Lm)ClpP2 is both structurally and functionally similar to previously studied tetradecameric ClpP proteins from Escherichia coli and Staphylococcus aureus, heptameric LmClpP1 features an asparagine in its catalytic triad. Mutation of this asparagine to aspartate increased the reactivity of the active site and led to the assembly of a tetradecameric complex. We analyzed the heterooligomeric complex of LmClpP1 and LmClpP2 via coexpression and subsequent labeling studies with natural product-derived probes. Notably, the LmClpP1 peptidase activity is stimulated 75-fold in the complex providing insights into heterooligomerization as a regulatory mechanism. Collectively, our data point toward different preferences for substrates and inhibitors of the two ClpP enzymes and highlight their structural and functional characteristics.
Conflict of interest statement
Conflict of interest statement: M. Gersch, M.P., M.D., and S.A.S. are named inventors on a patent application describing fluorogenic substrates suitable for ClpP activity measurements.
Figures




References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

