Global transcriptome profiles of Camellia sinensis during cold acclimation
- PMID: 23799877
- PMCID: PMC3701547
- DOI: 10.1186/1471-2164-14-415
Global transcriptome profiles of Camellia sinensis during cold acclimation
Abstract
Background: Tea is the most popular non-alcoholic health beverage in the world. The tea plant (Camellia sinensis (L.) O. Kuntze) needs to undergo a cold acclimation process to enhance its freezing tolerance in winter. Changes that occur at the molecular level in response to low temperatures are poorly understood in tea plants. To elucidate the molecular mechanisms of cold acclimation, we employed RNA-Seq and digital gene expression (DGE) technologies to the study of genome-wide expression profiles during cold acclimation in tea plants.
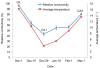
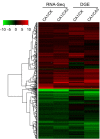
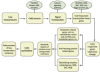
Results: Using the Illumina sequencing platform, we obtained approximately 57.35 million RNA-Seq reads. These reads were assembled into 216,831 transcripts, with an average length of 356 bp and an N50 of 529 bp. In total, 1,770 differentially expressed transcripts were identified, of which 1,168 were up-regulated and 602 down-regulated. These include a group of cold sensor or signal transduction genes, cold-responsive transcription factor genes, plasma membrane stabilization related genes, osmosensing-responsive genes, and detoxification enzyme genes. DGE and quantitative RT-PCR analysis further confirmed the results from RNA-Seq analysis. Pathway analysis indicated that the "carbohydrate metabolism pathway" and the "calcium signaling pathway" might play a vital role in tea plants' responses to cold stress.
Conclusions: Our study presents a global survey of transcriptome profiles of tea plants in response to low, non-freezing temperatures and yields insights into the molecular mechanisms of tea plants during the cold acclimation process. It could also serve as a valuable resource for relevant research on cold-tolerance and help to explore the cold-related genes in improving the understanding of low-temperature tolerance and plant-environment interactions.
Figures



References
-
- Hare PD, Cress WA, Van Staden J. Dissecting the roles of osmolyte accumulation during stress. Plant Cell Environ. 1998;21(6):535–553. doi: 10.1046/j.1365-3040.1998.00309.x. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

