Functional characterisation of the non-essential protein kinases and phosphatases regulating Aspergillus nidulans hydrolytic enzyme production
- PMID: 23800192
- PMCID: PMC3698209
- DOI: 10.1186/1754-6834-6-91
Functional characterisation of the non-essential protein kinases and phosphatases regulating Aspergillus nidulans hydrolytic enzyme production
Abstract
Background: Despite recent advances in the understanding of lignocellulolytic enzyme regulation, less is known about how different carbon sources are sensed and the signaling cascades that result in the adaptation of cellular metabolism and hydrolase secretion. Therefore, the role played by non-essential protein kinases (NPK) and phosphatases (NPP) in the sensing of carbon and/or energetic status was investigated in the model filamentous fungus Aspergillus nidulans.
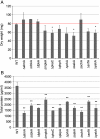
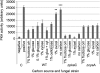
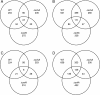
Results: Eleven NPKs and seven NPPs were identified as being involved in cellulase, and in some cases also hemicellulase, production in A. nidulans. The regulation of CreA-mediated carbon catabolite repression (CCR) in the parental strain was determined by fluorescence microscopy, utilising a CreA::GFP fusion protein. The sensing of phosphorylated glucose, via the RAS signalling pathway induced CreA repression, while carbon starvation resulted in derepression. Growth on cellulose represented carbon starvation and derepressing conditions. The involvement of the identified NPKs in the regulation of cellulose-induced responses and CreA derepression was assessed by genome-wide transcriptomics (GEO accession 47810). CreA::GFP localisation and the restoration of endocellulase activity via the introduction of the ∆creA mutation, was assessed in the NPK-deficient backgrounds. The absence of either the schA or snfA kinase dramatically reduced cellulose-induced transcriptional responses, including the expression of hydrolytic enzymes and transporters. The mechanism by which these two NPKs controlled gene transcription was identified, as the NPK-deficient mutants were not able to unlock CreA-mediated carbon catabolite repression under derepressing conditions, such as carbon starvation or growth on cellulose.
Conclusions: Collectively, this study identified multiple kinases and phosphatases involved in the sensing of carbon and/or energetic status, while demonstrating the overlapping, synergistic roles of schA and snfA in the regulation of CreA derepression and hydrolytic enzyme production in A. nidulans. The importance of a carbon starvation-induced signal for CreA derepression, permitting transcriptional activator binding, appeared paramount for hydrolase secretion.
Figures






References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

