A high-throughput microfluidic dental plaque biofilm system to visualize and quantify the effect of antimicrobials
- PMID: 23800904
- PMCID: PMC3797639
- DOI: 10.1093/jac/dkt211
A high-throughput microfluidic dental plaque biofilm system to visualize and quantify the effect of antimicrobials
Abstract
Objectives: Few model systems are amenable to developing multi-species biofilms in parallel under environmentally germane conditions. This is a problem when evaluating the potential real-world effectiveness of antimicrobials in the laboratory. One such antimicrobial is cetylpyridinium chloride (CPC), which is used in numerous over-the-counter oral healthcare products. The aim of this work was to develop a high-throughput microfluidic system that is combined with a confocal laser scanning microscope (CLSM) to quantitatively evaluate the effectiveness of CPC against oral multi-species biofilms grown in human saliva.
Methods: Twenty-four-channel BioFlux microfluidic plates were inoculated with pooled human saliva and fed filter-sterilized saliva for 20 h at 37°C. The bacterial diversity of the biofilms was evaluated by bacterial tag-encoded FLX amplicon pyrosequencing (bTEFAP). The antimicrobial/anti-biofilm effect of CPC (0.5%-0.001% w/v) was examined using Live/Dead stain, CLSM and 3D imaging software.
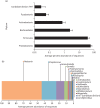
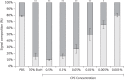
Results: The analysis of biofilms by bTEFAP demonstrated that they contained genera typically found in human dental plaque. These included Aggregatibacter, Fusobacterium, Neisseria, Porphyromonas, Streptococcus and Veillonella. Using Live/Dead stain, clear gradations in killing were observed when the biofilms were treated with CPC between 0.5% and 0.001% w/v. At 0.5% (w/v) CPC, 90% of the total signal was from dead/damaged cells. Below this concentration range, less killing was observed. In the 0.5%-0.05% (w/v) range CPC penetration/killing was greatest and biofilm thickness was significantly reduced.
Conclusions: This work demonstrates the utility of a high-throughput microfluidic-CLSM system to grow multi-species oral biofilms, which are compositionally similar to naturally occurring biofilms, to assess the effectiveness of antimicrobials.
Keywords: Live/Dead staining; confocal scanning laser microscopy; microfluidics; multi-species biofilm; pyrosequencing.
Figures



Similar articles
-
High-Velocity Microsprays Enhance Antimicrobial Activity in Streptococcus mutans Biofilms.J Dent Res. 2016 Dec;95(13):1494-1500. doi: 10.1177/0022034516662813. Epub 2016 Aug 23. J Dent Res. 2016. PMID: 27554642
-
L-arginine destabilizes oral multi-species biofilm communities developed in human saliva.PLoS One. 2015 May 6;10(5):e0121835. doi: 10.1371/journal.pone.0121835. eCollection 2015. PLoS One. 2015. PMID: 25946040 Free PMC article.
-
Characterization and application of a flow system for in vitro multispecies oral biofilm formation.J Periodontal Res. 2014 Jun;49(3):323-32. doi: 10.1111/jre.12110. Epub 2013 Jul 1. J Periodontal Res. 2014. PMID: 23815431
-
Multispecies communities: interspecies interactions influence growth on saliva as sole nutritional source.Int J Oral Sci. 2011 Apr;3(2):49-54. doi: 10.4248/IJOS11025. Int J Oral Sci. 2011. PMID: 21485308 Free PMC article. Review.
-
Saliva and dental plaque.Adv Dent Res. 2000 Dec;14:29-39. doi: 10.1177/08959374000140010401. Adv Dent Res. 2000. PMID: 11842921 Review.
Cited by
-
Interruption of Electrical Conductivity of Titanium Dental Implants Suggests a Path Towards Elimination Of Corrosion.PLoS One. 2015 Oct 13;10(10):e0140393. doi: 10.1371/journal.pone.0140393. eCollection 2015. PLoS One. 2015. PMID: 26461491 Free PMC article.
-
A Sensitive Thresholding Method for Confocal Laser Scanning Microscope Image Stacks of Microbial Biofilms.Sci Rep. 2018 Aug 29;8(1):13013. doi: 10.1038/s41598-018-31012-5. Sci Rep. 2018. PMID: 30158655 Free PMC article.
-
In vitro model systems for exploring oral biofilms: From single-species populations to complex multi-species communities.J Appl Microbiol. 2022 Feb;132(2):855-871. doi: 10.1111/jam.15200. Epub 2021 Sep 2. J Appl Microbiol. 2022. PMID: 34216534 Free PMC article. Review.
-
Microfluidic organ-on-chip systems for periodontal research: advances and future directions.Front Bioeng Biotechnol. 2025 Jan 7;12:1490453. doi: 10.3389/fbioe.2024.1490453. eCollection 2024. Front Bioeng Biotechnol. 2025. PMID: 39840127 Free PMC article. Review.
-
Dextranase enhances nanoparticle penetration of S. mutans biofilms.J Oral Microbiol. 2025 Jul 15;17(1):2528561. doi: 10.1080/20002297.2025.2528561. eCollection 2025. J Oral Microbiol. 2025. PMID: 40672866 Free PMC article.
References
-
- Costerton JW, Cheng KJ, Geesey GG, et al. Bacterial biofilms in nature and disease. Annu Rev Microbiol. 1987;41:435–64. - PubMed
-
- Stoodley P, Sauer K, Davies DG, et al. Biofilms as complex differentiated communities. Annu Rev Microbiol. 2002;56:187–209. - PubMed
-
- Armitage GC. Basic features of biofilms–why are they difficult therapeutic targets? Ann R Australas Coll Dent Surg. 2004;17:30–4. - PubMed
-
- Gilbert P, Maira-Litran T, McBain AJ, et al. The physiology and collective recalcitrance of microbial biofilm communities. Adv Microb Physiol. 2002;46:202–56. - PubMed
-
- Mah TF, O'Toole GA. Mechanisms of biofilm resistance to antimicrobial agents. Trends Microbiol. 2001;9:34–9. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

