Emerging antiviral strategies to interfere with influenza virus entry
- PMID: 23801557
- PMCID: PMC7168512
- DOI: 10.1002/med.21289
Emerging antiviral strategies to interfere with influenza virus entry
Abstract
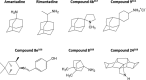
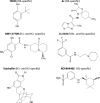
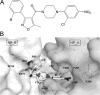
Influenza A and B viruses are highly contagious respiratory pathogens with a considerable medical and socioeconomical burden and known pandemic potential. Current influenza vaccines require annual updating and provide only partial protection in some risk groups. Due to the global spread of viruses with resistance to the M2 proton channel inhibitor amantadine or the neuraminidase inhibitor oseltamivir, novel antiviral agents with an original mode of action are urgently needed. We here focus on emerging options to interfere with the influenza virus entry process, which consists of the following steps: attachment of the viral hemagglutinin to the sialylated host cell receptors, endocytosis, M2-mediated uncoating, low pH-induced membrane fusion, and, finally, import of the viral ribonucleoprotein into the nucleus. We review the current functional and structural insights in the viral and cellular components of this entry process, and the diverse antiviral strategies that are being explored. This encompasses small molecule inhibitors as well as macromolecules such as therapeutic antibodies. There is optimism that at least some of these innovative concepts to block influenza virus entry will proceed from the proof of concept to a more advanced stage. Special attention is therefore given to the challenging issues of influenza virus (sub)type-dependent activity or potential drug resistance.
Keywords: M2 channel; antiviral; hemagglutinin; influenza virus; nucleoprotein.
© 2013 Wiley Periodicals, Inc.
Figures
Similar articles
-
Inhibitory and combinatorial effect of diphyllin, a v-ATPase blocker, on influenza viruses.Antiviral Res. 2013 Sep;99(3):371-82. doi: 10.1016/j.antiviral.2013.06.014. Epub 2013 Jun 29. Antiviral Res. 2013. PMID: 23820269 Free PMC article.
-
Identification of Clotrimazole Derivatives as Specific Inhibitors of Arenavirus Fusion.J Virol. 2019 Mar 5;93(6):e01744-18. doi: 10.1128/JVI.01744-18. Print 2019 Mar 15. J Virol. 2019. PMID: 30626681 Free PMC article.
-
Identification of pyrrolo[3,2-c]pyridin-4-amine compounds as a new class of entry inhibitors against influenza viruses in vitro.Biochem Biophys Res Commun. 2016 Sep 30;478(4):1594-601. doi: 10.1016/j.bbrc.2016.08.162. Epub 2016 Aug 29. Biochem Biophys Res Commun. 2016. PMID: 27586275
-
Perspective of Use of Antiviral Peptides against Influenza Virus.Viruses. 2015 Oct 20;7(10):5428-42. doi: 10.3390/v7102883. Viruses. 2015. PMID: 26492266 Free PMC article. Review.
-
Inhibitors targeting the influenza virus hemagglutinin.Curr Med Chem. 2015;22(11):1361-82. doi: 10.2174/0929867322666150227153919. Curr Med Chem. 2015. PMID: 25723505 Review.
Cited by
-
A small molecule compound targeting hemagglutinin inhibits influenza A virus and exhibits broad-spectrum antiviral activity.Acta Pharmacol Sin. 2024 Nov;45(11):2380-2393. doi: 10.1038/s41401-024-01331-7. Epub 2024 Jul 10. Acta Pharmacol Sin. 2024. PMID: 38987389
-
The pathogenesis of influenza in intact alveoli: virion endocytosis and its effects on the lung's air-blood barrier.Front Immunol. 2024 Jan 26;15:1328453. doi: 10.3389/fimmu.2024.1328453. eCollection 2024. Front Immunol. 2024. PMID: 38343548 Free PMC article. Review.
-
Novel Naturally Occurring Dipeptides and Single-Stranded Oligonucleotide Act as Entry Inhibitors and Exhibit a Strong Synergistic Anti-HIV-1 Profile.Infect Dis Ther. 2022 Jun;11(3):1103-1116. doi: 10.1007/s40121-022-00626-8. Epub 2022 Apr 7. Infect Dis Ther. 2022. PMID: 35391633 Free PMC article.
-
Analysis of the Genetic Diversity Associated With the Drug Resistance and Pathogenicity of Influenza A Virus Isolated in Bangladesh From 2002 to 2019.Front Microbiol. 2021 Sep 17;12:735305. doi: 10.3389/fmicb.2021.735305. eCollection 2021. Front Microbiol. 2021. PMID: 34603265 Free PMC article. Review.
-
Potential of acylated peptides to target the influenza A virus.Beilstein J Org Chem. 2015 Apr 29;11:589-95. doi: 10.3762/bjoc.11.65. eCollection 2015. Beilstein J Org Chem. 2015. PMID: 26124860 Free PMC article.
References
-
- Monto AS. Epidemiology of influenza. Vaccine 2008;26(Suppl 4):D45–8.:D45–D48. - PubMed
-
- Schanzer DL, Langley JM, Tam TW. Hospitalization attributable to influenza and other viral respiratory illnesses in Canadian children. Pediatr Infect Dis J 2006;25:795–800. - PubMed
-
- Thompson WW, Comanor L, Shay DK. Epidemiology of seasonal influenza: use of surveillance data and statistical models to estimate the burden of disease. J Infect Dis 2006;194(Suppl 2):S82–S91. - PubMed
-
- Cocoros N, Hernandez R, Harrington N, Rausch‐Phung E, Schulte CR, Blog D, Gallagher K, Klevos A, Kim C, Marano N, Alvarado‐Ramy. Estimates of deaths associated with seasonal influenza—United States, 1976–2007. MMWR Morb Mortal Wkly Rep 2010;59:1057–1062. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

