Perspectives on the RNA polymerase II core promoter
- PMID: 23801666
- PMCID: PMC3695423
- DOI: 10.1002/wdev.21
Perspectives on the RNA polymerase II core promoter
Abstract
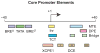
The RNA polymerase II core promoter is sometimes referred to as the gateway to transcription. The core promoter is generally defined to be the stretch of DNA that directs the initiation of transcription. This simple description belies a complex multidimensional regulatory element, as there is considerable diversity in core promoter structure and function. Core promoters can be viewed at the levels of DNA sequences, transcription factors, and biological networks. Key DNA sequences are known as core promoter elements, which include the TATA box, initiator (Inr), polypyrimidine initiator (TCT), TFIIB recognition element (BRE), motif ten element (MTE), and downstream core promoter element (DPE) motifs. There are no universal core promoter elements that are present in all promoters. Different types of core promoters are transcribed by different sets of transcription factors and exhibit distinct properties, such as specific interactions with transcriptional enhancers, that are determined by the presence or absence of particular core promoter motifs. Moreover, some core promoter elements have been found to be associated with specific biological networks. For instance, the TCT motif is dedicated to the transcription of ribosomal protein genes in Drosophila and humans. In addition, nearly all of the Drosophila Hox genes have a DPE motif in their core promoters. The complexity of the core promoter is further seen in the relation among transcription initiation patterns, the stability or lability of transcriptional states, and the organization of the chromatin structure in the promoter region. Hence, the current data indicate that the core promoter is a critical component in the regulation of gene activity.
Copyright © 2011 Wiley Periodicals, Inc.
Figures

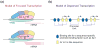

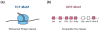
References
-
- Smale ST, Kadonaga JT. The RNA polymerase II core promoter. Annu Rev Biochem. 2003;72:449–479. - PubMed
-
- Sandelin A, Carninci P, Lenhard B, Ponjavic J, Hayashizaki Y, Hume DA. Mammalian RNA polymerase II core promoters: insights from genome-wide studies. Nat Rev Genet. 2007;8:424–436. - PubMed
-
- Thomas MC, Chiang CM. The general transcription machinery and general cofactors. Crit Rev Biochem Mol Biol. 2006;41:105–178. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

