Trinucleotide repeats: a structural perspective
- PMID: 23801983
- PMCID: PMC3687200
- DOI: 10.3389/fneur.2013.00076
Trinucleotide repeats: a structural perspective
Abstract
Trinucleotide repeat (TNR) expansions are present in a wide range of genes involved in several neurological disorders, being directly involved in the molecular mechanisms underlying pathogenesis through modulation of gene expression and/or the function of the RNA or protein it encodes. Structural and functional information on the role of TNR sequences in RNA and protein is crucial to understand the effect of TNR expansions in neurodegeneration. Therefore, this review intends to provide to the reader a structural and functional view of TNR and encoded homopeptide expansions, with a particular emphasis on polyQ expansions and its role at inducing the self-assembly, aggregation and functional alterations of the carrier protein, which culminates in neuronal toxicity and cell death. Detail will be given to the Machado-Joseph Disease-causative and polyQ-containing protein, ataxin-3, providing clues for the impact of polyQ expansion and its flanking regions in the modulation of ataxin-3 molecular interactions, function, and aggregation.
Keywords: amino acid-repeats; amyloid; microsatellites; protein aggregation; protein complexes; protein structure.
Figures

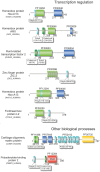


References
LinkOut - more resources
Full Text Sources
Other Literature Sources

