Detailed analysis of putative genes encoding small proteins in legume genomes
- PMID: 23802007
- PMCID: PMC3687714
- DOI: 10.3389/fpls.2013.00208
Detailed analysis of putative genes encoding small proteins in legume genomes
Abstract
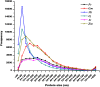
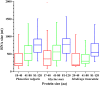
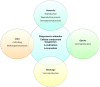
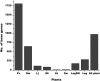
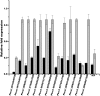
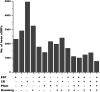
Diverse plant genome sequencing projects coupled with powerful bioinformatics tools have facilitated massive data analysis to construct specialized databases classified according to cellular function. However, there are still a considerable number of genes encoding proteins whose function has not yet been characterized. Included in this category are small proteins (SPs, 30-150 amino acids) encoded by short open reading frames (sORFs). SPs play important roles in plant physiology, growth, and development. Unfortunately, protocols focused on the genome-wide identification and characterization of sORFs are scarce or remain poorly implemented. As a result, these genes are underrepresented in many genome annotations. In this work, we exploited publicly available genome sequences of Phaseolus vulgaris, Medicago truncatula, Glycine max, and Lotus japonicus to analyze the abundance of annotated SPs in plant legumes. Our strategy to uncover bona fide sORFs at the genome level was centered in bioinformatics analysis of characteristics such as evidence of expression (transcription), presence of known protein regions or domains, and identification of orthologous genes in the genomes explored. We collected 6170, 10,461, 30,521, and 23,599 putative sORFs from P. vulgaris, G. max, M. truncatula, and L. japonicus genomes, respectively. Expressed sequence tags (ESTs) available in the DFCI Gene Index database provided evidence that ~one-third of the predicted legume sORFs are expressed. Most potential SPs have a counterpart in a different plant species and counterpart regions or domains in larger proteins. Potential functional sORFs were also classified according to a reduced set of GO categories, and the expression of 13 of them during P. vulgaris nodule ontogeny was confirmed by qPCR. This analysis provides a collection of sORFs that potentially encode for meaningful SPs, and offers the possibility of their further functional evaluation.
Keywords: gene annotation; legume genomes; short open reading frames.
Figures


Similar articles
-
Small open reading frames in plant research: from prediction to functional characterization.3 Biotech. 2022 Mar;12(3):76. doi: 10.1007/s13205-022-03147-w. Epub 2022 Feb 24. 3 Biotech. 2022. PMID: 35251879 Free PMC article. Review.
-
Computational discovery and annotation of conserved small open reading frames in fungal genomes.BMC Bioinformatics. 2019 Feb 4;19(Suppl 13):551. doi: 10.1186/s12859-018-2550-2. BMC Bioinformatics. 2019. PMID: 30717662 Free PMC article.
-
Discovery and annotation of small proteins using genomics, proteomics, and computational approaches.Genome Res. 2011 Apr;21(4):634-41. doi: 10.1101/gr.109280.110. Epub 2011 Mar 2. Genome Res. 2011. PMID: 21367939 Free PMC article.
-
Genome-wide analysis of the basic leucine zipper (bZIP) transcription factor gene family in six legume genomes.BMC Genomics. 2015 Dec 10;16:1053. doi: 10.1186/s12864-015-2258-x. BMC Genomics. 2015. PMID: 26651343 Free PMC article.
-
The Emerging World of Small ORFs.Trends Plant Sci. 2016 Apr;21(4):317-328. doi: 10.1016/j.tplants.2015.11.005. Epub 2015 Dec 10. Trends Plant Sci. 2016. PMID: 26684391 Review.
Cited by
-
Pervasive translation of small open reading frames in plant long non-coding RNAs.Front Plant Sci. 2022 Oct 24;13:975938. doi: 10.3389/fpls.2022.975938. eCollection 2022. Front Plant Sci. 2022. PMID: 36352887 Free PMC article. Review.
-
Genetic Diversity, Nitrogen Fixation, and Water Use Efficiency in a Panel of Honduran Common Bean (Phaseolus vulgaris L.) Landraces and Modern Genotypes.Plants (Basel). 2020 Sep 19;9(9):1238. doi: 10.3390/plants9091238. Plants (Basel). 2020. PMID: 32961677 Free PMC article.
-
Emerging evidence for functional peptides encoded by short open reading frames.Nat Rev Genet. 2014 Mar;15(3):193-204. doi: 10.1038/nrg3520. Epub 2014 Feb 11. Nat Rev Genet. 2014. PMID: 24514441 Review.
-
Beyond traditional translation: ncRNA derived peptides as modulators of tumor behaviors.J Biomed Sci. 2024 Jun 14;31(1):63. doi: 10.1186/s12929-024-01047-0. J Biomed Sci. 2024. PMID: 38877495 Free PMC article. Review.
-
Small open reading frames in plant research: from prediction to functional characterization.3 Biotech. 2022 Mar;12(3):76. doi: 10.1007/s13205-022-03147-w. Epub 2022 Feb 24. 3 Biotech. 2022. PMID: 35251879 Free PMC article. Review.
References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

