Mutations affecting G-protein subunit α11 in hypercalcemia and hypocalcemia
- PMID: 23802516
- PMCID: PMC3773604
- DOI: 10.1056/NEJMoa1300253
Mutations affecting G-protein subunit α11 in hypercalcemia and hypocalcemia
Abstract
Background: Familial hypocalciuric hypercalcemia is a genetically heterogeneous disorder with three variants: types 1, 2, and 3. Type 1 is due to loss-of-function mutations of the calcium-sensing receptor, a guanine nucleotide-binding protein (G-protein)-coupled receptor that signals through the G-protein subunit α11 (Gα11). Type 3 is associated with adaptor-related protein complex 2, sigma 1 subunit (AP2S1) mutations, which result in altered calcium-sensing receptor endocytosis. We hypothesized that type 2 is due to mutations effecting Gα11 loss of function, since Gα11 is involved in calcium-sensing receptor signaling, and its gene (GNA11) and the type 2 locus are colocalized on chromosome 19p13.3. We also postulated that mutations effecting Gα11 gain of function, like the mutations effecting calcium-sensing receptor gain of function that cause autosomal dominant hypocalcemia type 1, may lead to hypocalcemia.
Methods: We performed GNA11 mutational analysis in a kindred with familial hypocalciuric hypercalcemia type 2 and in nine unrelated patients with familial hypocalciuric hypercalcemia who did not have mutations in the gene encoding the calcium-sensing receptor (CASR) or AP2S1. We also performed this analysis in eight unrelated patients with hypocalcemia who did not have CASR mutations. In addition, we studied the effects of GNA11 mutations on Gα11 protein structure and calcium-sensing receptor signaling in human embryonic kidney 293 (HEK293) cells.
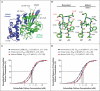
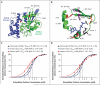
Results: The kindred with familial hypocalciuric hypercalcemia type 2 had an in-frame deletion of a conserved Gα11 isoleucine (Ile200del), and one of the nine unrelated patients with familial hypocalciuric hypercalcemia had a missense GNA11 mutation (Leu135Gln). Missense GNA11 mutations (Arg181Gln and Phe341Leu) were detected in two unrelated patients with hypocalcemia; they were therefore identified as having autosomal dominant hypocalcemia type 2. All four GNA11 mutations predicted disrupted protein structures, and assessment on the basis of in vitro expression showed that familial hypocalciuric hypercalcemia type 2-associated mutations decreased the sensitivity of cells expressing calcium-sensing receptors to changes in extracellular calcium concentrations, whereas autosomal dominant hypocalcemia type 2-associated mutations increased cell sensitivity.
Conclusions: Gα11 mutants with loss of function cause familial hypocalciuric hypercalcemia type 2, and Gα11 mutants with gain of function cause a clinical disorder designated as autosomal dominant hypocalcemia type 2. (Funded by the United Kingdom Medical Research Council and others.).
Figures


Comment in
-
G proteins--the disease spectrum expands.N Engl J Med. 2013 Jun 27;368(26):2515-6. doi: 10.1056/NEJMe1304751. N Engl J Med. 2013. PMID: 23802519 No abstract available.
References
-
- Hannan FM, Nesbit MA, Zhang C, et al. Identification of 70 calcium-sensing receptor mutations in hyper- and hypocalcaemic patients: evidence for clustering of extracellular domain mutations at calcium-binding sites. Hum Mol Genet. 2012;21:2768–78. - PubMed
-
- Pollak MR, Brown EM, Chou YH, et al. Mutations in the human Ca(2+)-sensing receptor gene cause familial hypocalciuric hypercalcemia and neonatal severe hyperparathyroidism. Cell. 1993;75:1297–303. - PubMed
-
- Christensen SE, Nissen PH, Vestergaard P, et al. Plasma 25-hydroxyvitamin D, 1,25-dihydroxyvitamin D, and parathyroid hormone in familial hypocalciuric hypercalcemia and primary hyperpara thyroidism. Eur J Endocrinol. 2008;159:719–27. - PubMed
-
- Thakker RV. Diseases associated with the extracellular calcium-sensing receptor. Cell Calcium. 2004;35:275–82. - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
