Intact cluster and chordate-like expression of ParaHox genes in a sea star
- PMID: 23803323
- PMCID: PMC3710244
- DOI: 10.1186/1741-7007-11-68
Intact cluster and chordate-like expression of ParaHox genes in a sea star
Abstract
Background: The ParaHox genes are thought to be major players in patterning the gut of several bilaterian taxa. Though this is a fundamental role that these transcription factors play, their activities are not limited to the endoderm and extend to both ectodermal and mesodermal tissues. Three genes compose the ParaHox group: Gsx, Xlox and Cdx. In some taxa (mostly chordates but to some degree also in protostomes) the three genes are arranged into a genomic cluster, in a similar fashion to what has been shown for the better-known Hox genes. Sea urchins possess the full complement of ParaHox genes but they are all dispersed throughout the genome, an arrangement that, perhaps, represented the primitive condition for all echinoderms. In order to understand the evolutionary history of this group of genes we cloned and characterized all ParaHox genes, studied their expression patterns and identified their genomic loci in a member of an earlier branching group of echinoderms, the asteroid Patiria miniata.
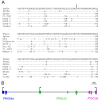
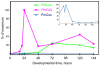
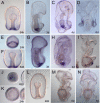
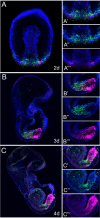
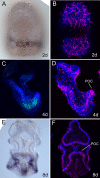
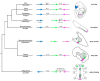
Results: We identified the three ParaHox orthologs in the genome of P. miniata. While one of them, PmGsx is provided as maternal message, with no zygotic activation afterwards, the other two, PmLox and PmCdx are expressed during embryogenesis, within restricted domains of both endoderm and ectoderm. Screening of a Patiria bacterial artificial chromosome (BAC) library led to the identification of a clone containing the three genes. The transcriptional directions of PmGsx and PmLox are opposed to that of the PmCdx gene within the cluster.
Conclusions: The identification of P. miniata ParaHox genes has revealed the fact that these genes are clustered in the genome, in contrast to what has been reported for echinoids. Since the presence of an intact cluster, or at least a partial cluster, has been reported in chordates and polychaetes respectively, it becomes clear that within echinoderms, sea urchins have modified the original bilaterian arrangement. Moreover, the sea star ParaHox domains of expression show chordate-like features not found in the sea urchin, confirming that the dynamics of gene expression for the respective genes and their putative regulatory interactions have clearly changed over evolutionary time within the echinoid lineage.
Figures
Similar articles
-
Genetic organization and embryonic expression of the ParaHox genes in the sea urchin S. purpuratus: insights into the relationship between clustering and colinearity.Dev Biol. 2006 Dec 1;300(1):63-73. doi: 10.1016/j.ydbio.2006.07.037. Epub 2006 Aug 4. Dev Biol. 2006. PMID: 16959236
-
Identification of an intact ParaHox cluster with temporal colinearity but altered spatial colinearity in the hemichordate Ptychodera flava.BMC Evol Biol. 2013 Jun 27;13:129. doi: 10.1186/1471-2148-13-129. BMC Evol Biol. 2013. PMID: 23802544 Free PMC article.
-
Minimal ProtoHox cluster inferred from bilaterian and cnidarian Hox complements.Nature. 2006 Aug 10;442(7103):684-7. doi: 10.1038/nature04863. Nature. 2006. PMID: 16900199
-
Time is of the essence for ParaHox homeobox gene clustering.BMC Biol. 2013 Jun 26;11:72. doi: 10.1186/1741-7007-11-72. BMC Biol. 2013. PMID: 23803337 Free PMC article. Review.
-
Evolution of invertebrate deuterostomes and Hox/ParaHox genes.Genomics Proteomics Bioinformatics. 2011 Jun;9(3):77-96. doi: 10.1016/S1672-0229(11)60011-9. Genomics Proteomics Bioinformatics. 2011. PMID: 21802045 Free PMC article. Review.
Cited by
-
Systematic comparison of sea urchin and sea star developmental gene regulatory networks explains how novelty is incorporated in early development.Nat Commun. 2020 Dec 4;11(1):6235. doi: 10.1038/s41467-020-20023-4. Nat Commun. 2020. PMID: 33277483 Free PMC article.
-
A pancreatic exocrine-like cell regulatory circuit operating in the upper stomach of the sea urchin Strongylocentrotus purpuratus larva.BMC Evol Biol. 2016 May 26;16(1):117. doi: 10.1186/s12862-016-0686-0. BMC Evol Biol. 2016. PMID: 27230062 Free PMC article.
-
Transcriptomic analysis of sea star development through metamorphosis to the highly derived pentameral body plan with a focus on neural transcription factors.DNA Res. 2020 Feb 1;27(1):dsaa007. doi: 10.1093/dnares/dsaa007. DNA Res. 2020. PMID: 32339242 Free PMC article.
-
The ParaHox gene Gsx patterns the apical organ and central nervous system but not the foregut in scaphopod and cephalopod mollusks.Evodevo. 2015 Dec 29;6:41. doi: 10.1186/s13227-015-0037-z. eCollection 2015. Evodevo. 2015. PMID: 26715985 Free PMC article.
-
Non-collinear Hox gene expression in bivalves and the evolution of morphological novelties in mollusks.Sci Rep. 2021 Feb 11;11(1):3575. doi: 10.1038/s41598-021-82122-6. Sci Rep. 2021. PMID: 33574385 Free PMC article.
References
-
- Arenas-Mena C, Cameron AR, Davidson EH. Spatial expression of Hox cluster genes in the ontogeny of a sea urchin. Development. 2000;127:4631–4643. - PubMed
-
- Seo HC, Edvardsen RB, Maeland AD, Bjordal M, Jensen MF, Hansen A, Flaat M, Weissenbach J, Lehrach H, Wincker P, Reinhardt R, Chourrout D. Hox cluster disintegration with persistent anteroposterior order of expression in Oikopleura dioica. Nature. 2004;431:67–71. doi: 10.1038/nature02709. - DOI - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

